A data compendium associating the genomes of 12,289 Mycobacterium tuberculosis isolates with quantitative resistance phenotypes to 13 antibiotics
- PMID: 35944069
- PMCID: PMC9363010
- DOI: 10.1371/journal.pbio.3001721
A data compendium associating the genomes of 12,289 Mycobacterium tuberculosis isolates with quantitative resistance phenotypes to 13 antibiotics
Abstract
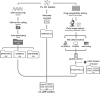
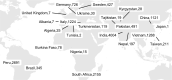
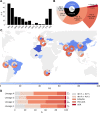
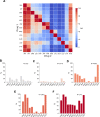
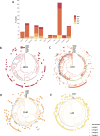
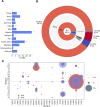
The Comprehensive Resistance Prediction for Tuberculosis: an International Consortium (CRyPTIC) presents here a data compendium of 12,289 Mycobacterium tuberculosis global clinical isolates, all of which have undergone whole-genome sequencing and have had their minimum inhibitory concentrations to 13 antitubercular drugs measured in a single assay. It is the largest matched phenotypic and genotypic dataset for M. tuberculosis to date. Here, we provide a summary detailing the breadth of data collected, along with a description of how the isolates were selected, collected, and uniformly processed in CRyPTIC partner laboratories across 23 countries. The compendium contains 6,814 isolates resistant to at least 1 drug, including 2,129 samples that fully satisfy the clinical definitions of rifampicin resistant (RR), multidrug resistant (MDR), pre-extensively drug resistant (pre-XDR), or extensively drug resistant (XDR). The data are enriched for rare resistance-associated variants, and the current limits of genotypic prediction of resistance status (sensitive/resistant) are presented by using a genetic mutation catalogue, along with the presence of suspected resistance-conferring mutations for isolates resistant to the newly introduced drugs bedaquiline, clofazimine, delamanid, and linezolid. Finally, a case study of rifampicin monoresistance demonstrates how this compendium could be used to advance our genetic understanding of rare resistance phenotypes. The data compendium is fully open source and it is hoped that it will facilitate and inspire future research for years to come.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: E.R. is employed by Public Health England and holds an honorary contract with Imperial College London. I.F.L. is Director of the Scottish Mycobacteria Reference Laboratory. S.N. receives funding from German Center for Infection Research, Excellenz Cluster Precision Medicine in Chronic Inflammation, Leibniz Science Campus Evolutionary Medicine of the LUNG (EvoLUNG)tion EXC 2167. P.S. is a consultant at Genoscreen. T.R. is funded by NIH and DoD and receives salary support from the non-profit organization FIND. T.R. is a co-founder, board member and shareholder of Verus Diagnostics Inc, a company that was founded with the intent of developing diagnostic assays. Verus Diagnostics was not involved in any way with data collection, analysis or publication of the results. T.R. has not received any financial support from Verus Diagnostics. UCSD Conflict of Interest office has reviewed and approved T.R.’s role in Verus Diagnostics Inc. T.R. is a co-inventor of a provisional patent for a TB diagnostic assay (provisional patent #: 63/048.989). T.R. is a co-inventor on a patent associated with the processing of TB sequencing data (European Patent Application No. 14840432.0 & USSN 14/912,918). T.R. has agreed to “donate all present and future interest in and rights to royalties from this patent” to UCSD to ensure that he does not receive any financial benefits from this patent. S.S. is working and holding ESOPs at HaystackAnalytics Pvt. Ltd. (Product: Using whole genome sequencing for drug susceptibility testing for Mycobacterium tuberculosis). G.F.G. is listed as an inventor on patent applications for RBD-dimer-based CoV vaccines. The patents for RBD-dimers as protein subunit vaccines for SARS-CoV-2 have been licensed to Anhui Zhifei Longcom Biopharmaceutical Co. Ltd, China.
Figures
References
-
- WHO. Global Tuberculosis Report 2020. 2021.
-
- World Health Organisation. EndTB Campaign. Available from: www.who.int/teams/global-tuberculosis-programme/the-end-tb-strategy.
-
- Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U60 OE000103/OE/OSELS CDC HHS/United States
- 214560/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- T32 GM007365/GM/NIGMS NIH HHS/United States
- 203135/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 098316 /WT_/Wellcome Trust/United Kingdom
- 203141/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI124413/AI/NIAID NIH HHS/United States
- MRC_/Medical Research Council/United Kingdom
- 206724/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI146338/AI/NIAID NIH HHS/United States
- 201470/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 101237/Z/13/B/WT_/Wellcome Trust/United Kingdom
- 214321/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- 200205/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 203583/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- BRC-1215-20008/DH_/Department of Health/United Kingdom
- FC0010218/WT_/Wellcome Trust/United Kingdom
- R01 AI114900/AI/NIAID NIH HHS/United States
- R03 AI117312/AI/NIAID NIH HHS/United States
- FC0010218/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

