Pan-cancer integrative histology-genomic analysis via multimodal deep learning
- PMID: 35944502
- PMCID: PMC10397370
- DOI: 10.1016/j.ccell.2022.07.004
Pan-cancer integrative histology-genomic analysis via multimodal deep learning
Abstract
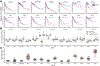
The rapidly emerging field of computational pathology has demonstrated promise in developing objective prognostic models from histology images. However, most prognostic models are either based on histology or genomics alone and do not address how these data sources can be integrated to develop joint image-omic prognostic models. Additionally, identifying explainable morphological and molecular descriptors from these models that govern such prognosis is of interest. We use multimodal deep learning to jointly examine pathology whole-slide images and molecular profile data from 14 cancer types. Our weakly supervised, multimodal deep-learning algorithm is able to fuse these heterogeneous modalities to predict outcomes and discover prognostic features that correlate with poor and favorable outcomes. We present all analyses for morphological and molecular correlates of patient prognosis across the 14 cancer types at both a disease and a patient level in an interactive open-access database to allow for further exploration, biomarker discovery, and feature assessment.
Keywords: artificial intelligence; biomarker discovery; cancer prognosis; computational pathology; data fusion; deep learning; digital pathology; multimodal integration; multimodal prognostic models; pan-cancer.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.J.C. and F.M. are inventors on a patent that has been filed corresponding multimodal data fusion using deep learning. The authors declare no other competing interests.
Figures





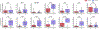
Comment in
-
Human and machine: Better at pathology together?Cancer Cell. 2022 Aug 8;40(8):806-808. doi: 10.1016/j.ccell.2022.06.004. Cancer Cell. 2022. PMID: 35944500
-
Multimodal deep learning: An improvement in prognostication or a reflection of batch effect?Cancer Cell. 2023 Jan 9;41(1):5-6. doi: 10.1016/j.ccell.2022.10.025. Epub 2022 Nov 10. Cancer Cell. 2023. PMID: 36368319 No abstract available.
References
-
- Abdelmoula WM, Balluff B, Englert S, Dijkstra J, Reinders MJ, Walch A, McDonnell LA and Lelieveldt BP, (2016). Data-driven identification of prognostic tumor subpopulations using spatially mapped t-SNE of mass spectrometry imaging data. Proceedings of the National Academy of Sciences, 113(43), pp.12244–12249. 10.1073/pnas.1510227113 - DOI - PMC - PubMed
-
- AbdulJabbar K, Raza SEA, Rosenthal R, Jamal-Hanjani M, Veeriah S, Akarca A, Lund T, Moore DA, Salgado R, Al Bakir M. and Zapata L, (2020). Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nature Medicine, 26(7), pp.1054–1062. 10.1038/s41591-020-0900-x - DOI - PMC - PubMed
-
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and Winchester DP, (2017). The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population‐ based to a more “personalized” approach to cancer staging. CA: a cancer journal for clinicians, 67(2), pp.93–99. 10.3322/caac.21388 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

