Molecular heterogeneity and commonalities in pancreatic cancer precursors with gastric and intestinal phenotype
- PMID: 35944927
- PMCID: PMC9933174
- DOI: 10.1136/gutjnl-2021-326550
Molecular heterogeneity and commonalities in pancreatic cancer precursors with gastric and intestinal phenotype
Abstract
Objective: Due to the limited number of modifiable risk factors, secondary prevention strategies based on early diagnosis represent the preferred route to improve the prognosis of pancreatic ductal adenocarcinoma (PDAC). Here, we provide a comparative morphogenetic analysis of PDAC precursors aiming at dissecting the process of carcinogenesis and tackling the heterogeneity of preinvasive lesions.
Design: Targeted and whole-genome low-coverage sequencing, genome-wide methylation and transcriptome analyses were applied on a final collective of 122 morphologically well-characterised low-grade and high-grade PDAC precursors, including intestinal and gastric intraductal papillary mucinous neoplasms (IPMN) and pancreatic intraepithelial neoplasias (PanIN).
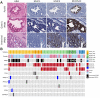
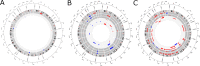
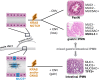
Results: Epigenetic regulation of mucin genes determines the phenotype of PDAC precursors. PanIN and gastric IPMN display a ductal molecular profile and numerous similarly regulated pathways, including the Notch pathway, but can be distinguished by recurrent deletions and differential methylation and, in part, by the expression of mucin-like 3. Intestinal IPMN are clearly distinct lesions at the molecular level with a more instable genotype and are possibly related to a different ductal cell compartment.
Conclusions: PDAC precursors with gastric and intestinal phenotype are heterogeneous in terms of morphology, genetic and epigenetic profile. This heterogeneity is related to a different cell identity and, possibly, to a different aetiology.
Keywords: gene expression; gene mutation; pancreatic pathology; pancreatic tumours; pre-malignancy - GI tract.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
