Drosophila Homolog of the Human Carpenter Syndrome Linked Gene, MEGF8, Is Required for Synapse Development and Function
- PMID: 35944997
- PMCID: PMC9480877
- DOI: 10.1523/JNEUROSCI.0442-22.2022
Drosophila Homolog of the Human Carpenter Syndrome Linked Gene, MEGF8, Is Required for Synapse Development and Function
Abstract
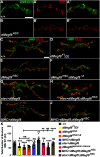
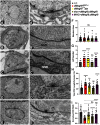
Drosophila multiple epidermal growth factor-like domains 8 (dMegf8) is a homolog of human MEGF8 MEGF8 encodes a multidomain transmembrane protein which is highly conserved across species. In humans, MEGF8 mutations cause a rare genetic disorder called Carpenter syndrome, which is frequently associated with abnormal left-right patterning, cardiac defects, and learning disabilities. MEGF8 is also associated with psychiatric disorders. Despite its clinical relevance, MEGF8 remains poorly characterized; and although it is highly conserved, studies on animal models of Megf8 are also very limited. The presence of intellectual disabilities in Carpenter syndrome patients and association of MEGF8 with psychiatric disorders indicate that mutations in MEGF8 cause underlying defects in synaptic structure and functions. In this study, we investigated the role of Drosophila dMegf8 in glutamatergic synapses of the larval neuromuscular junctions (NMJ) in both males and females. We show that dMegf8 localizes to NMJ synapses and is required for proper synaptic growth. dMegf8 mutant larvae and adults show severe motor coordination deficits. At the NMJ, dMegf8 mutants show altered localization of presynaptic and postsynaptic proteins, defects in synaptic ultrastructure, and neurotransmission. Interestingly, dMegf8 mutants have reduced levels of the Type II BMP receptor Wishful thinking (Wit). dMegf8 displays genetic interactions with neurexin-1 (dnrx) and wit, and in association with Dnrx and Wit plays an essential role in synapse organization. Our studies provide insights into human MEGF8 functions and potentially into mechanisms that may underlie intellectual disabilities observed in Carpenter syndrome as well as MEGF8-related synaptic structural and/or functional deficits in psychiatric disorders.SIGNIFICANCE STATEMENT Carpenter syndrome, known for over a century now, is a genetic disorder linked to mutations in Multiple Epidermal Growth Factor-like Domains 8 (MEGF8) gene and associated with intellectual disabilities among other symptoms. MEGF8 is also associated with psychiatric disorders. Despite the high genetic conservation and clinical relevance, the functions of MEGF8 remain largely uncharacterized. Patients with intellectual disabilities and psychiatric diseases often have an underlying defect in synaptic structure and function. This work defines the role of the fly homolog of human MEGF8, dMegf8, in glutamatergic synapse growth, organization, and function and provide insights into potential functions of MEGF8 in human central synapses and synaptic mechanisms that may underlie psychiatric disorders and intellectual disabilities seen in Carpenter syndrome.
Keywords: BMP signaling; Carpenter syndrome; Drosophila larval NMJ; MEGF8; Neurexin-1; synapses.
Copyright © 2022 the authors.
Figures







Similar articles
-
Neurexin, Neuroligin and Wishful Thinking coordinate synaptic cytoarchitecture and growth at neuromuscular junctions.Mol Cell Neurosci. 2017 Jan;78:9-24. doi: 10.1016/j.mcn.2016.11.004. Epub 2016 Nov 10. Mol Cell Neurosci. 2017. PMID: 27838296 Free PMC article.
-
Mutations in multidomain protein MEGF8 identify a Carpenter syndrome subtype associated with defective lateralization.Am J Hum Genet. 2012 Nov 2;91(5):897-905. doi: 10.1016/j.ajhg.2012.08.027. Epub 2012 Oct 11. Am J Hum Genet. 2012. PMID: 23063620 Free PMC article.
-
Coordinated Regulation of Axonal Microtubule Organization and Transport by Drosophila Neurexin and BMP Pathway.Sci Rep. 2018 Nov 26;8(1):17337. doi: 10.1038/s41598-018-35618-7. Sci Rep. 2018. PMID: 30478335 Free PMC article.
-
Synapse development and maturation at the drosophila neuromuscular junction.Neural Dev. 2020 Aug 2;15(1):11. doi: 10.1186/s13064-020-00147-5. Neural Dev. 2020. PMID: 32741370 Free PMC article. Review.
-
Development and plasticity of the Drosophila larval neuromuscular junction.Wiley Interdiscip Rev Dev Biol. 2013 Sep-Oct;2(5):647-70. doi: 10.1002/wdev.108. Epub 2013 Feb 5. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 24014452 Free PMC article. Review.
Cited by
-
Immunohistochemical and Immunoelectron Microscopical Distribution of MEGF8 in the Mouse Central Nervous System.Cells. 2023 Dec 28;13(1):63. doi: 10.3390/cells13010063. Cells. 2023. PMID: 38201267 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
