A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia
- PMID: 35945197
- PMCID: PMC9363439
- DOI: 10.1038/s41467-022-31860-w
A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia
Abstract
Emerging diseases caused by coronaviruses of likely bat origin (e.g., SARS, MERS, SADS, COVID-19) have disrupted global health and economies for two decades. Evidence suggests that some bat SARS-related coronaviruses (SARSr-CoVs) could infect people directly, and that their spillover is more frequent than previously recognized. Each zoonotic spillover of a novel virus represents an opportunity for evolutionary adaptation and further spread; therefore, quantifying the extent of this spillover may help target prevention programs. We derive current range distributions for known bat SARSr-CoV hosts and quantify their overlap with human populations. We then use probabilistic risk assessment and data on human-bat contact, human viral seroprevalence, and antibody duration to estimate that a median of 66,280 people (95% CI: 65,351-67,131) are infected with SARSr-CoVs annually in Southeast Asia. These data on the geography and scale of spillover can be used to target surveillance and prevention programs for potential future bat-CoV emergence.
© 2022. The Author(s).
Conflict of interest statement
P.D. served as a member of the WHO-China joint study on COVID-19 origins, is a current member of the Taskforce on the Origins and Early Spread of COVID-19 and One Health Solutions to Future Pandemics, and was Chair of the IPBES Pandemics and Biodiversity Workshop. P.D. has made numerous public statements both independently, and as part of these groups, on the likely origins of COVID-19 and the need to assess risk and prioritize targeted surveillance for future disease emergence. L.F.W. serves on multiple committees for WHO, FAO, and OIE on COVID-19 including assay and vaccine development and animal models; has ongoing research investigating the origin of COVID-19; and has made statements on this issue to the media. P.Z. serves on multiple committees for MOST (China) on COVID-19 including pathogen identification, monitoring viral mutations, and the likely origin of COVID-19. Z.L.S. has made statements to the media about the likely origins of COVID-19. The remaining authors declare no competing interests.
Figures
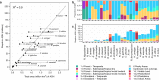
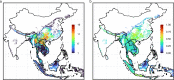
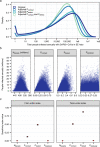

Update of
-
A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia.medRxiv [Preprint]. 2021 Sep 14:2021.09.09.21263359. doi: 10.1101/2021.09.09.21263359. medRxiv. 2021. Update in: Nat Commun. 2022 Aug 9;13(1):4380. doi: 10.1038/s41467-022-31860-w. PMID: 34545371 Free PMC article. Updated. Preprint.
Comment in
-
Tens of thousands of people exposed to bat coronaviruses each year.Nature. 2022 Aug;608(7923):457-458. doi: 10.1038/d41586-022-02153-5. Nature. 2022. PMID: 35945279 No abstract available.
Similar articles
-
A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia.medRxiv [Preprint]. 2021 Sep 14:2021.09.09.21263359. doi: 10.1101/2021.09.09.21263359. medRxiv. 2021. Update in: Nat Commun. 2022 Aug 9;13(1):4380. doi: 10.1038/s41467-022-31860-w. PMID: 34545371 Free PMC article. Updated. Preprint.
-
Strategy To Assess Zoonotic Potential Reveals Low Risk Posed by SARS-Related Coronaviruses from Bat and Pangolin.mBio. 2023 Apr 25;14(2):e0328522. doi: 10.1128/mbio.03285-22. Epub 2023 Feb 14. mBio. 2023. PMID: 36786573 Free PMC article.
-
Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China.mBio. 2022 Jun 28;13(3):e0046322. doi: 10.1128/mbio.00463-22. Epub 2022 Apr 25. mBio. 2022. PMID: 35467426 Free PMC article.
-
Ecology, evolution and spillover of coronaviruses from bats.Nat Rev Microbiol. 2022 May;20(5):299-314. doi: 10.1038/s41579-021-00652-2. Epub 2021 Nov 19. Nat Rev Microbiol. 2022. PMID: 34799704 Free PMC article. Review.
-
Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
Cited by
-
Neo-humanism and COVID-19: Opportunities for a socially and environmentally sustainable world.Appl Res Qual Life. 2023;18(1):9-41. doi: 10.1007/s11482-022-10112-5. Epub 2022 Dec 9. Appl Res Qual Life. 2023. PMID: 36530493 Free PMC article.
-
International Collaboration is the Only Way to Protect Ourselves from the Next Pandemic.Ecohealth. 2022 Sep;19(3):317-319. doi: 10.1007/s10393-022-01609-4. Epub 2022 Aug 8. Ecohealth. 2022. PMID: 35939132 Free PMC article. No abstract available.
-
Present and future distribution of bat hosts of sarbecoviruses: implications for conservation and public health.Proc Biol Sci. 2022 May 25;289(1975):20220397. doi: 10.1098/rspb.2022.0397. Epub 2022 May 25. Proc Biol Sci. 2022. PMID: 35611534 Free PMC article.
-
Dynamics of natural selection preceding human viral epidemics and pandemics.bioRxiv [Preprint]. 2025 Feb 28:2025.02.26.640439. doi: 10.1101/2025.02.26.640439. bioRxiv. 2025. PMID: 40060453 Free PMC article. Preprint.
-
Building a pathway to One Health surveillance and response in Asian countries.Sci One Health. 2024 Apr 2;3:100067. doi: 10.1016/j.soh.2024.100067. eCollection 2024. Sci One Health. 2024. PMID: 39077383 Free PMC article. Review.
References
-
- Lee J-W, McKibbin WJ. Globalization and disease: the case of SARS. Asian Economic Pap. 2004;3:113–131. doi: 10.1162/1535351041747932. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

