Repeated genetic adaptation to altitude in two tropical butterflies
- PMID: 35945236
- PMCID: PMC9363431
- DOI: 10.1038/s41467-022-32316-x
Repeated genetic adaptation to altitude in two tropical butterflies
Abstract
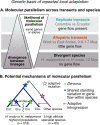
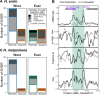
Repeated evolution can provide insight into the mechanisms that facilitate adaptation to novel or changing environments. Here we study adaptation to altitude in two tropical butterflies, Heliconius erato and H. melpomene, which have repeatedly and independently adapted to montane habitats on either side of the Andes. We sequenced 518 whole genomes from altitudinal transects and found many regions differentiated between highland (~ 1200 m) and lowland (~ 200 m) populations. We show repeated genetic differentiation across replicate populations within species, including allopatric comparisons. In contrast, there is little molecular parallelism between the two species. By sampling five close relatives, we find that a large proportion of divergent regions identified within species have arisen from standing variation and putative adaptive introgression from high-altitude specialist species. Taken together our study supports a role for both standing genetic variation and gene flow from independently adapted species in promoting parallel local adaptation to the environment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Martin A, Orgogozo V. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

