Construction of a hypoxia-derived gene model to predict the prognosis and therapeutic response of head and neck squamous cell carcinoma
- PMID: 35945448
- PMCID: PMC9363468
- DOI: 10.1038/s41598-022-17898-2
Construction of a hypoxia-derived gene model to predict the prognosis and therapeutic response of head and neck squamous cell carcinoma
Abstract
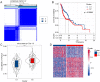
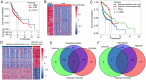
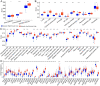
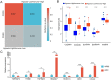
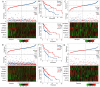
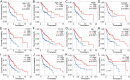
Head and neck squamous cell carcinoma (HNSCC) ranks as the sixth most common cancer worldwide and has a poor prognosis in the advanced stage. Increasing evidence has shown that hypoxia contributes to genetic alterations that have essential effects on the occurrence and progression of cancers. However, the exact roles hypoxia-related genes play in HNSCC remain unclear. In this study, we downloaded the mRNA expression profiles and clinical data of patients with HNSCC from The Cancer Genome Atlas and Gene Expression Omnibus. Two molecular subtypes were identified based on prognostic hypoxia-related genes using the ConsensusClusterPlus method. ESTIMATE was used to calculate the immune score of each patient. Kyoto Encyclopedia of Genes and Genomes and Gene Ontology were used for functional annotation. A prognostic risk model was generated by Cox regression and least absolute shrinkage and selection operator analysis. We identified two distinct molecular subtypes, cluster 1 and cluster 2, based on 200 hypoxia-related genes. Additionally, we identified three hypoxia-immune subgroups (hypoxia-high/immune-low, hypoxia-low/immune-high, and mixed subgroups). The hypoxia-high/immune-low group had the worst prognosis, while the hypoxia-low/immune-high group had the best prognosis. Patients in the hypoxia-low/immune-high group were more sensitive to anti-PD-L1 treatment and chemotherapy than those in the hypoxia-high/immune-low group. Furthermore, we constructed a prognostic risk model based on the differentially expressed genes between the hypoxia-immune subgroups. The survival analysis and time-dependent ROC analysis results demonstrated the good performance of the established 7-gene signature for predicting HNSCC prognosis. In conclusions, the constructed hypoxia-related model might serve as a promising biomarker for the diagnosis and prognosis of HNSCC, and it could predict immunotherapy and chemotherapy efficacy in HNSCC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

