Genome-wide identification and expression analysis of carotenoid cleavage oxygenase genes in Litchi (Litchi chinensis Sonn.)
- PMID: 35945492
- PMCID: PMC9361530
- DOI: 10.1186/s12870-022-03772-w
Genome-wide identification and expression analysis of carotenoid cleavage oxygenase genes in Litchi (Litchi chinensis Sonn.)
Abstract
Background: Carotenoid cleavage oxygenases (CCOs) include the carotenoid cleavage dioxygenase (CCD) and 9-cis-epoxycarotenoid (NCED), which can catalize carotenoid to form various apocarotenoids and their derivatives, has been found that play important role in the plant world. But little information of CCO gene family has been reported in litchi (Litchi chinensis Sonn.) till date.
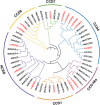
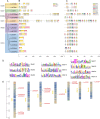
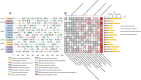
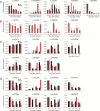
Results: In this study, a total of 15 LcCCO genes in litchi were identified based on genome wide lever. Phylogeny analysis showed that LcCCO genes could be classified into six subfamilies (CCD1, CCD4, CCD7, CCD8, CCD-like, and NCED), which gene structure, domain and motifs exhibited similar distribution patterns in the same subfamilies. MiRNA target site prediction found that there were 32 miRNA target sites in 13 (86.7%) LcCCO genes. Cis-elements analysis showed that the largest groups of elements were light response related, following was plant hormones, stress and plant development related. Expression pattern analysis revealed that LcCCD4, LcNCED1, and LcNCED2 might be involving with peel coloration, LcCCDlike-b might be an important factor deciding fruit flavor, LcNCED2 and LcNCED3 might be related to flower control, LcNCED1 and LcNCED2 might function in fruitlet abscission, LcCCD4a1, LcCCD4a2, LcCCD1, LcCCD4, LcNCED1, and LcNCED2 might participate in postharvest storage of litchi.
Conclusion: Herein, Genome-wide analysis of the LcCCO genes was conducted in litchi to investigate their structure features and potential functions. These valuable and expectable information of LcCCO genes supplying in this study will offer further more possibility to promote quality improvement and breeding of litchi and further function investigation of this gene family in plant.
Keywords: CCO genes; Expression analysis; Flower control; Fruit development and maturation; Litchi; Postharvest storage.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Genome-Wide Identification and Expression Analysis of m6A Writers, Erasers, and Readers in Litchi (Litchi chinensis Sonn.).Genes (Basel). 2022 Dec 4;13(12):2284. doi: 10.3390/genes13122284. Genes (Basel). 2022. PMID: 36553551 Free PMC article.
-
Genome-wide analysis and identification of Carotenoid Cleavage Oxygenase (CCO) gene family in coffee (coffee arabica) under abiotic stress.BMC Genom Data. 2024 Jul 19;25(1):71. doi: 10.1186/s12863-024-01248-4. BMC Genom Data. 2024. PMID: 39030545 Free PMC article.
-
Genome-wide analysis of carotenoid cleavage oxygenases and identification of ripening-associated DzNCED5a in durian (Durio zibethinus) fruit.Plant Physiol Biochem. 2024 Jan;206:108253. doi: 10.1016/j.plaphy.2023.108253. Epub 2023 Dec 3. Plant Physiol Biochem. 2024. PMID: 38086212
-
Plant carotenoid cleavage oxygenases: structure-function relationships and role in development and metabolism.Brief Funct Genomics. 2020 Jan 22;19(1):1-9. doi: 10.1093/bfgp/elz037. Brief Funct Genomics. 2020. PMID: 31875900 Review.
-
Sugar Transport, Metabolism and Signaling in Fruit Development of Litchi chinensis Sonn: A Review.Int J Mol Sci. 2021 Oct 18;22(20):11231. doi: 10.3390/ijms222011231. Int J Mol Sci. 2021. PMID: 34681891 Free PMC article. Review.
Cited by
-
SiNCED1, a 9-cis-epoxycarotenoid dioxygenase gene in Setaria italica, is involved in drought tolerance and seed germination in transgenic Arabidopsis.Front Plant Sci. 2023 Mar 9;14:1121809. doi: 10.3389/fpls.2023.1121809. eCollection 2023. Front Plant Sci. 2023. PMID: 36968367 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of Carotenoid Cleavage Dioxygenase Genes in Salvia miltiorrhiza.Int J Mol Sci. 2024 Dec 6;25(23):13138. doi: 10.3390/ijms252313138. Int J Mol Sci. 2024. PMID: 39684848 Free PMC article.
-
Genome-wide identification and in-silico expression analysis of carotenoid cleavage oxygenases gene family in Oryza sativa (rice) in response to abiotic stress.Front Plant Sci. 2023 Oct 25;14:1269995. doi: 10.3389/fpls.2023.1269995. eCollection 2023. Front Plant Sci. 2023. PMID: 37954992 Free PMC article.
-
Genome-wide identification and in-silico expression analysis of CCO gene family in sunflower (Helianthus annnus) against abiotic stress.Plant Mol Biol. 2024 Apr 3;114(2):34. doi: 10.1007/s11103-024-01433-0. Plant Mol Biol. 2024. PMID: 38568355 Free PMC article.
-
Genome-wide identification and characterization of NCED gene family in soybean (Glycine max L.) and their expression profiles in response to various abiotic stress treatments.PLoS One. 2025 Mar 25;20(3):e0319952. doi: 10.1371/journal.pone.0319952. eCollection 2025. PLoS One. 2025. PMID: 40131870 Free PMC article.
References
-
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, et al. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006;45(6):982–993. doi: 10.1111/j.1365-313X.2006.02666.x. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 2019RC082/Natural Science Foundation of Hainan Province
- 2019RC082/Natural Science Foundation of Hainan Province
- 2019RC082/Natural Science Foundation of Hainan Province
- 2019RC082/Natural Science Foundation of Hainan Province
- KYQD(2R)1970/Initial funding Hainan University introduces high-level personnel
- KYQD(2R)1970/Initial funding Hainan University introduces high-level personnel
- KYQD(2R)1970/Initial funding Hainan University introduces high-level personnel
- KYQD(2R)1970/Initial funding Hainan University introduces high-level personnel
- 31960570/National Natural Science Foundation of China
- 31960570/National Natural Science Foundation of China
- 31960570/National Natural Science Foundation of China
- 32060655/National Natural Science Foundation of China
- 32060655/National Natural Science Foundation of China
- 31960570/National Natural Science Foundation of China
- 31960570/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

