Balanced mitochondrial and cytosolic translatomes underlie the biogenesis of human respiratory complexes
- PMID: 35945592
- PMCID: PMC9361522
- DOI: 10.1186/s13059-022-02732-9
Balanced mitochondrial and cytosolic translatomes underlie the biogenesis of human respiratory complexes
Abstract
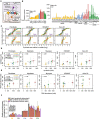
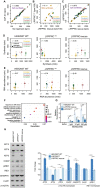
Background: Oxidative phosphorylation (OXPHOS) complexes consist of nuclear and mitochondrial DNA-encoded subunits. Their biogenesis requires cross-compartment gene regulation to mitigate the accumulation of disproportionate subunits. To determine how human cells coordinate mitochondrial and nuclear gene expression processes, we tailored ribosome profiling for the unique features of the human mitoribosome.
Results: We resolve features of mitochondrial translation initiation and identify a small ORF in the 3' UTR of MT-ND5. Analysis of ribosome footprints in five cell types reveals that average mitochondrial synthesis levels correspond precisely to cytosolic levels across OXPHOS complexes, and these average rates reflect the relative abundances of the complexes. Balanced mitochondrial and cytosolic synthesis does not rely on rapid feedback between the two translation systems, and imbalance caused by mitochondrial translation deficiency is associated with the induction of proteotoxicity pathways.
Conclusions: Based on our findings, we propose that human OXPHOS complexes are synthesized proportionally to each other, with mitonuclear balance relying on the regulation of OXPHOS subunit translation across cellular compartments, which may represent a proteostasis vulnerability.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

