A framework for testing the impact of co-infections on host gut microbiomes
- PMID: 35945629
- PMCID: PMC9361228
- DOI: 10.1186/s42523-022-00198-5
A framework for testing the impact of co-infections on host gut microbiomes
Abstract
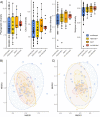
Parasitic infections disturb gut microbial communities beyond their natural range of variation, possibly leading to dysbiosis. Yet it remains underappreciated that most infections are accompanied by one or more co-infections and their collective impact is largely unexplored. Here we developed a framework illustrating changes to the host gut microbiome following single infections, and build on it by describing the neutral, synergistic or antagonistic impacts on microbial α- and ß-diversity expected from co-infections. We tested the framework on microbiome data from a non-human primate population co-infected with helminths and Adenovirus, and matched patterns reported in published studies to the introduced framework. In this case study, α-diversity of co-infected Malagasy mouse lemurs (Microcebus griseorufus) did not differ in comparison with that of singly infected or uninfected individuals, even though community composition captured with ß-diversity metrices changed significantly. Explicitly, we record stochastic changes in dispersion, a sign of dysbiosis, following the Anna-Karenina principle rather than deterministic shifts in the microbial gut community. From the literature review and our case study, neutral and synergistic impacts emerged as common outcomes from co-infections, wherein both shifts and dispersion of microbial communities following co-infections were often more severe than after a single infection alone, but microbial α-diversity was not universally altered. Important functions of the microbiome may also suffer from such heavily altered, though no less species-rich microbial community. Lastly, we pose the hypothesis that the reshuffling of host-associated microbial communities due to the impact of various, often coinciding parasitic infections may become a source of novel or zoonotic diseases.
Keywords: Co-infections; Disease ecology; Dysbiosis; Gut microbiome; Helminths; Non-human primate; One health; Parasites; Virus; Wildlife health.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
