Functional compensation of mouse duplicates by their paralogs expressed in the same tissues
- PMID: 35945673
- PMCID: PMC9387915
- DOI: 10.1093/gbe/evac126
Functional compensation of mouse duplicates by their paralogs expressed in the same tissues
Abstract
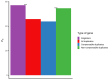
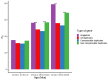
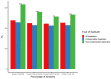
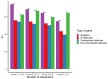
Analyses in a number of organisms have shown that duplicated genes are less likely to be essential than singletons. This implies that genes can often compensate for the loss of their paralogs. However, it is unclear why the loss of some duplicates can be compensated by their paralogs, whereas the loss of other duplicates cannot. Surprisingly, initial analyses in mice did not detect differences in the essentiality of duplicates and singletons. Only subsequent analyses, using larger gene knockout datasets and controlling for a number of confounding factors, did detect significant differences. Previous studies have not taken into account the tissues in which duplicates are expressed. We hypothesized that in complex organisms, in order for a gene's loss to be compensated by one or more of its paralogs, such paralogs need to be expressed in at least the same set of tissues as the lost gene. To test our hypothesis, we classified mouse duplicates into two categories based on the expression patterns of their paralogs: "compensable duplicates" (those with paralogs expressed in all the tissues in which the gene is expressed) and "non-compensable duplicates" (those whose paralogs are not expressed in all the tissues where the gene is expressed). In agreement with our hypothesis, the essentiality of non-compensable duplicates is similar to that of singletons, whereas compensable duplicates exhibit a substantially lower essentiality. Our results imply that duplicates can often compensate for the loss of their paralogs, but only if they are expressed in the same tissues. Indeed, the compensation ability is more dependent on expression patterns than on protein sequence similarity. The existence of these two kinds of duplicates with different essentialities, which has been overlooked by prior studies, may have hindered the detection of differences between singletons and duplicates.
Keywords: Duplicates; essentiality; gene expression; singletons.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
References
-
- Barau J, et al. 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354:909–912. - PubMed
LinkOut - more resources
Full Text Sources

