Zanthoxylum bungeanum root-rot associated shifts in microbiomes of root endosphere, rhizosphere, and soil
- PMID: 35945942
- PMCID: PMC9357373
- DOI: 10.7717/peerj.13808
Zanthoxylum bungeanum root-rot associated shifts in microbiomes of root endosphere, rhizosphere, and soil
Abstract
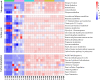
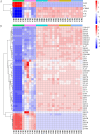
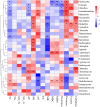
Root-rot disease has lead to serious reduction in yields and jeopardized the survival of the economically and ecologically important Zanthoxylum bungeanum trees cultured in Sichuan Province. In order to investigate the interaction between the microbiome and the root-rot disease, a metagenomic analysis was performed to characterize the microbial communities and functions in Z. bungeanum root endosphere, rhizosphere and bulk soil with/without root-rot disease. Soil physicochemical properties, microbial population size and enzyme activities were also analyzed for finding their interactions with the root-rot disease. As results, lower total nitrogen (TN) and available phosphorus (AP) contents but higher pH in rhizosphere and bulk soil, as well as lower substrate-induced respiration (SIR) and higher protease activity in bulk soil of diseased trees were found, in comparison with that of healthy trees. Microbial diversity and community composition were changed by root-rot disease in the endosphere, but not in rhizosphere and bulk soils. The endophytic microbiome of diseased trees presented higher Proteobacteria abundance and lower abundances of Bacteroidetes, Firmicutes and dominant fungal phyla. The relative abundances of nitrogen cycle- and carbon cycle-related genes in endophytic microbiomes were different between the diseased and healthy trees. Based on ANOSIM and PCoA, functional profiles (KEGG and CAZy) of microbiomes in rhizosphere and bulk soil shifted significantly between the diseased and healthy trees. In addition, soil pH, TN, AP, SIR, invertase and protease were estimated as the main factors influencing the shifts of taxonomic and functional groups in microbiomes of rhizosphere and bulk soil. Conclusively, the imbalance of root and soil microbial function groups might lead to shifts in the root endosphere-rhizosphere microenvironment, which in turn resulted in Z. bungeanum root-rot.
Keywords: Microbiome; Rhizosphere; Root endosphere; Root-rot diseases; Zanthoxylum bungeanum.
©2022 Liao et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures




References
-
- Bodah ET. Root rot diseases in plants: a review of common causal agents and management strategies. Agricultural Research & Technology. 2017;5:555661. doi: 10.19080/ARTOAJ.2017.05.555661. - DOI
-
- Bodenhausen N, Somerville V, Desiro A, Walser J-C, Borghi L, van der Heijden MG, Schlaeppi K. Petunia-and Arabidopsis-specific root microbiota responses to phosphate supplementation. Phytobiomes Journal. 2019;3:112–124. doi: 10.1094/PBIOMES-12-18-0057-R. - DOI
-
- Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecological Monographs. 1957;27:325–349.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

