Identification of Pathologic Grading-Related Genes Associated with Kidney Renal Clear Cell Carcinoma
- PMID: 35945960
- PMCID: PMC9357261
- DOI: 10.1155/2022/2818777
Identification of Pathologic Grading-Related Genes Associated with Kidney Renal Clear Cell Carcinoma
Abstract
Background: Renal epithelium lesions can cause renal cell carcinoma. This kind of tumor is common among all renal cancers with poor prognosis, of which more than 70% belong to kidney renal clear cell carcinoma. As the pathogenesis of KIRC has not been elucidated, it is necessary to be further explored.
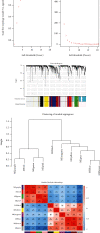
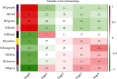
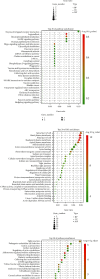
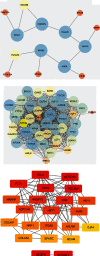
Methods: The Genomic Spatial Event database was used to obtain the analysis dataset (GSE126964) based on the GEO database, and The Cancer Genome Atlas was applied for KIRC data collection. edgeR and limma analyses were subsequently conducted to identify differentially expressed genes. Based on the systems biology approach of WGCNA, potential biomarkers and therapeutic targets of this disease were screened after the establishment of a gene coexpression network. GO and KEGG enrichment used cluster Profiler, enrichplot, and ggplot2 in the R software package. Protein-protein interaction network diagrams were plotted for hub gene collection via the STRING platform and Cytoscape software. Hub genes associated with overall survival time of KIRC patients were ultimately identified using the Kaplan-Meier plotter.
Results: There were 1863 DEGs identified in total and ten coexpressed gene modules discovered using a WGCNA method. GO and KEGG analysis findings revealed that the most enrichment pathways included Notch binding, cell migration, cell cycle, cell senescence, apoptosis, focal adhesions, and autophagosomes. Twenty-seven hub genes were identified, among which FLT1, HNRNPU, ATP6V0D2, ATP6V1A, and ATP6V1H were positively correlated with OS rates of KIRC patients (p < 0.05).
Conclusions: In conclusion, bioinformatic techniques can be useful tools for predicting the progression of KIRC. DEGs are present in both KIRC and normal kidney tissues, which can be considered the KIRC biomarkers.
Copyright © 2022 Weijian Xiong et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

