Evolution of the nitric oxide synthase family in vertebrates and novel insights in gill development
- PMID: 35946155
- PMCID: PMC9363997
- DOI: 10.1098/rspb.2022.0667
Evolution of the nitric oxide synthase family in vertebrates and novel insights in gill development
Abstract
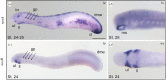
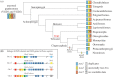
Nitric oxide (NO) is an ancestral key signalling molecule essential for life and has enormous versatility in biological systems, including cardiovascular homeostasis, neurotransmission and immunity. Although our knowledge of NO synthases (Nos), the enzymes that synthesize NO in vivo, is substantial, the origin of a large and diversified repertoire of nos gene orthologues in fishes with respect to tetrapods remains a puzzle. The recent identification of nos3 in the ray-finned fish spotted gar, which was considered lost in this lineage, changed this perspective. This finding prompted us to explore nos gene evolution, surveying vertebrate species representing key evolutionary nodes. This study provides noteworthy findings: first, nos2 experienced several lineage-specific gene duplications and losses. Second, nos3 was found to be lost independently in two different teleost lineages, Elopomorpha and Clupeocephala. Third, the expression of at least one nos paralogue in the gills of developing shark, bichir, sturgeon, and gar, but not in lamprey, suggests that nos expression in this organ may have arisen in the last common ancestor of gnathostomes. These results provide a framework for continuing research on nos genes' roles, highlighting subfunctionalization and reciprocal loss of function that occurred in different lineages during vertebrate genome duplications.
Keywords: gene duplication and loss; genome duplication; nos; phylogenomics; synteny; vertebrate evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Molecular evolution of nitric oxide synthases in metazoans.Comp Biochem Physiol Part D Genomics Proteomics. 2010 Dec;5(4):295-301. doi: 10.1016/j.cbd.2010.08.004. Epub 2010 Sep 9. Comp Biochem Physiol Part D Genomics Proteomics. 2010. PMID: 20884305
-
Tetrapod V1R-like ora genes in an early-diverging ray-finned fish species: the canonical six ora gene repertoire of teleost fish resulted from gene loss in a larger ancestral repertoire.BMC Genomics. 2016 Jan 27;17:83. doi: 10.1186/s12864-016-2399-6. BMC Genomics. 2016. PMID: 26818853 Free PMC article.
-
Dynamic evolution of transient receptor potential vanilloid (TRPV) ion channel family with numerous gene duplications and losses.Front Endocrinol (Lausanne). 2022 Nov 1;13:1013868. doi: 10.3389/fendo.2022.1013868. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36387917 Free PMC article.
-
New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts.Front Endocrinol (Lausanne). 2020 Sep 24;11:538196. doi: 10.3389/fendo.2020.538196. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33071966 Free PMC article. Review.
-
From 2R to 3R: evidence for a fish-specific genome duplication (FSGD).Bioessays. 2005 Sep;27(9):937-45. doi: 10.1002/bies.20293. Bioessays. 2005. PMID: 16108068 Review.
Cited by
-
Expression Pattern of nos1 in the Developing Nervous System of Ray-Finned Fish.Genes (Basel). 2022 May 20;13(5):918. doi: 10.3390/genes13050918. Genes (Basel). 2022. PMID: 35627303 Free PMC article.
-
Nitric Oxide Production and Regulation in the Teleost Cardiovascular System.Antioxidants (Basel). 2022 May 12;11(5):957. doi: 10.3390/antiox11050957. Antioxidants (Basel). 2022. PMID: 35624821 Free PMC article. Review.
-
Bone Formation in Zebrafish: The Significance of DAF-FM DA Staining for Nitric Oxide Detection.Biomolecules. 2023 Dec 12;13(12):1780. doi: 10.3390/biom13121780. Biomolecules. 2023. PMID: 38136650 Free PMC article.
-
Detecting Intestinal Goblet Cells of the Broadgilled Hagfish Eptatretus cirrhatus (Forster, 1801): A Confocal Microscopy Evaluation.Biology (Basel). 2022 Sep 17;11(9):1366. doi: 10.3390/biology11091366. Biology (Basel). 2022. PMID: 36138844 Free PMC article.
-
Nitric Oxide Function and Nitric Oxide Synthase Evolution in Aquatic Chordates.Int J Mol Sci. 2023 Jul 6;24(13):11182. doi: 10.3390/ijms241311182. Int J Mol Sci. 2023. PMID: 37446358 Free PMC article. Review.

