Multi-omics analysis for potential inflammation-related genes involved in tumour immune evasion via extended application of epigenetic data
- PMID: 35946310
- PMCID: PMC9364145
- DOI: 10.1098/rsob.210375
Multi-omics analysis for potential inflammation-related genes involved in tumour immune evasion via extended application of epigenetic data
Abstract
Accumulating evidence suggests that inflammation-related genes may play key roles in tumour immune evasion. Programmed cell death ligand 1 (PD-L1) is an important immune checkpoint involved in mediating anti-tumour immunity. We performed multi-omics analysis to explore key inflammation-related genes affecting the transcriptional regulation of PD-L1 expression. The open chromatin region of the PD-L1 promoter was mapped using the assay for transposase-accessible chromatin using sequencing (ATAC-seq) profiles. Correlation analysis of epigenetic data (ATAC-seq) and transcriptome data (RNA-seq) were performed to identify inflammation-related transcription factors (TFs) whose expression levels were correlated with the chromatin accessibility of the PD-L1 promoter. Chromatin immunoprecipitation sequencing (ChIP-seq) profiles were used to confirm the physical binding of the TF STAT2 and the predicted binding regions. We also confirmed the results of the bioinformatics analysis with cell experiments. We identified chr9 : 5449463-5449962 and chr9 : 5450250-5450749 as reproducible open chromatin regions in the PD-L1 promoter. Moreover, we observed a correlation between STAT2 expression and the accessibility of the aforementioned regions. Furthermore, we confirmed its physical binding through ChIP-seq profiles and demonstrated the regulation of PD-L1 by STAT2 overexpression in vitro. Multiple databases were also used for the validation of the results. Our study identified STAT2 as a direct upstream TF regulating PD-L1 expression. The interaction of STAT2 and PD-L1 might be associated with tumour immune evasion in cancers, suggesting the potential value for tumour treatment.
Keywords: ATAC-seq; PD-L1; STAT2; epigenetics; immune checkpoint therapy; immune evasion.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
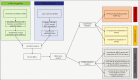

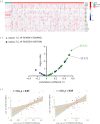
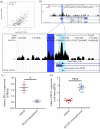

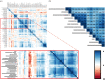
References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

