A genetic model for in vivo proximity labelling of the mammalian secretome
- PMID: 35946312
- PMCID: PMC9364151
- DOI: 10.1098/rsob.220149
A genetic model for in vivo proximity labelling of the mammalian secretome
Abstract
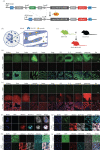
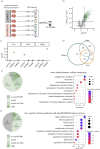
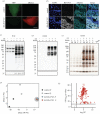
Organ functions are highly specialized and interdependent. Secreted factors regulate organ development and mediate homeostasis through serum trafficking and inter-organ communication. Enzyme-catalysed proximity labelling enables the identification of proteins within a specific cellular compartment. Here, we report a BirA*G3 mouse strain that enables CRE-dependent promiscuous biotinylation of proteins trafficking through the endoplasmic reticulum. When broadly activated throughout the mouse, widespread labelling of proteins was observed within the secretory pathway. Streptavidin affinity purification and peptide mapping by quantitative mass spectrometry (MS) proteomics revealed organ-specific secretory profiles and serum trafficking. As expected, secretory proteomes were highly enriched for signal peptide-containing proteins, highlighting both conventional and non-conventional secretory processes, and ectodomain shedding. Lower-abundance proteins with hormone-like properties were recovered and validated using orthogonal approaches. Hepatocyte-specific activation of BirA*G3 highlighted liver-specific biotinylated secretome profiles. The BirA*G3 mouse model demonstrates enhanced labelling efficiency and tissue specificity over viral transduction approaches and will facilitate a deeper understanding of secretory protein interplay in development, and in healthy and diseased adult states.
Keywords: BirA; TurboID; inter-organ communication; proximity-labelling; secretome; serum proteins.
Conflict of interest statement
A.P.M. is a scientific adviser for Novartis, TRESTLE BioTherapeutics, eGENESIS and IVIVA Medical. S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, Seer and PrognomIQ. A.Y.T. declares intellectual property interests around proximity labelling technology.
Figures



References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

