Evolutionary Changes in the Chromatin Landscape Contribute to Reorganization of a Developmental Gene Network During Rapid Life History Evolution in Sea Urchins
- PMID: 35946348
- PMCID: PMC9435058
- DOI: 10.1093/molbev/msac172
Evolutionary Changes in the Chromatin Landscape Contribute to Reorganization of a Developmental Gene Network During Rapid Life History Evolution in Sea Urchins
Abstract
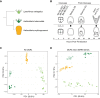
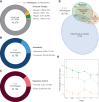
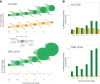
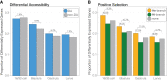
Chromatin configuration is highly dynamic during embryonic development in animals, exerting an important point of control in transcriptional regulation. Yet there exists remarkably little information about the role of evolutionary changes in chromatin configuration to the evolution of gene expression and organismal traits. Genome-wide assays of chromatin configuration, coupled with whole-genome alignments, can help address this gap in knowledge in several ways. In this study we present a comparative analysis of regulatory element sequences and accessibility throughout embryogenesis in three sea urchin species with divergent life histories: a lecithotroph Heliocidaris erythrogramma, a closely related planktotroph H. tuberculata, and a distantly related planktotroph Lytechinus variegatus. We identified distinct epigenetic and mutational signatures of evolutionary modifications to the function of putative cis-regulatory elements in H. erythrogramma that have accumulated nonuniformly throughout the genome, suggesting selection, rather than drift, underlies many modifications associated with the derived life history. Specifically, regulatory elements composing the sea urchin developmental gene regulatory network are enriched for signatures of positive selection and accessibility changes which may function to alter binding affinity and access of developmental transcription factors to these sites. Furthermore, regulatory element changes often correlate with divergent expression patterns of genes involved in cell type specification, morphogenesis, and development of other derived traits, suggesting these evolutionary modifications have been consequential for phenotypic evolution in H. erythrogramma. Collectively, our results demonstrate that selective pressures imposed by changes in developmental life history rapidly reshape the cis-regulatory landscape of core developmental genes to generate novel traits and embryonic programs.
Keywords: ATAC-seq; Evolution; development; gene regulation; life history; sea urchins.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




Similar articles
-
Single-Cell Transcriptomics Reveals Evolutionary Reconfiguration of Embryonic Cell Fate Specification in the Sea Urchin Heliocidaris erythrogramma.Genome Biol Evol. 2025 Jan 6;17(1):evae258. doi: 10.1093/gbe/evae258. Genome Biol Evol. 2025. PMID: 39587400 Free PMC article.
-
Comparative Developmental Transcriptomics Reveals Rewiring of a Highly Conserved Gene Regulatory Network during a Major Life History Switch in the Sea Urchin Genus Heliocidaris.PLoS Biol. 2016 Mar 4;14(3):e1002391. doi: 10.1371/journal.pbio.1002391. eCollection 2016 Mar. PLoS Biol. 2016. PMID: 26943850 Free PMC article.
-
Hybrid Epigenomes Reveal Extensive Local Genetic Changes to Chromatin Accessibility Contribute to Divergence in Embryonic Gene Expression Between Species.Mol Biol Evol. 2023 Nov 3;40(11):msad222. doi: 10.1093/molbev/msad222. Mol Biol Evol. 2023. PMID: 37823438 Free PMC article.
-
Morphological evolution in sea urchin development: hybrids provide insights into the pace of evolution.Bioessays. 2004 Apr;26(4):343-7. doi: 10.1002/bies.20024. Bioessays. 2004. PMID: 15057932 Review.
-
Experimentally based sea urchin gene regulatory network and the causal explanation of developmental phenomenology.Wiley Interdiscip Rev Syst Biol Med. 2009 Sep-Oct;1(2):237-246. doi: 10.1002/wsbm.24. Wiley Interdiscip Rev Syst Biol Med. 2009. PMID: 20228891 Free PMC article. Review.
Cited by
-
Recent reconfiguration of an ancient developmental gene regulatory network in Heliocidaris sea urchins.Nat Ecol Evol. 2022 Dec;6(12):1907-1920. doi: 10.1038/s41559-022-01906-9. Epub 2022 Oct 20. Nat Ecol Evol. 2022. PMID: 36266460
-
Single-cell transcriptomics reveals evolutionary reconfiguration of embryonic cell fate specification in the sea urchin Heliocidaris erythrogramma.bioRxiv [Preprint]. 2024 May 1:2024.04.30.591752. doi: 10.1101/2024.04.30.591752. bioRxiv. 2024. Update in: Genome Biol Evol. 2025 Jan 6;17(1):evae258. doi: 10.1093/gbe/evae258. PMID: 38746376 Free PMC article. Updated. Preprint.
-
Single-Cell Transcriptomics Reveals Evolutionary Reconfiguration of Embryonic Cell Fate Specification in the Sea Urchin Heliocidaris erythrogramma.Genome Biol Evol. 2025 Jan 6;17(1):evae258. doi: 10.1093/gbe/evae258. Genome Biol Evol. 2025. PMID: 39587400 Free PMC article.
-
Genome evolution and divergence in cis-regulatory architecture is associated with condition-responsive development in horned dung beetles.PLoS Genet. 2024 Mar 5;20(3):e1011165. doi: 10.1371/journal.pgen.1011165. eCollection 2024 Mar. PLoS Genet. 2024. PMID: 38442113 Free PMC article.
-
Chromatin profiling data indicate regulatory mechanisms for differentiation during development in the acoel Hofstenia miamia.Development. 2025 May 15;152(10):dev204799. doi: 10.1242/dev.204799. Epub 2025 May 23. Development. 2025. PMID: 40326510
References
-
- Byrne M, Sewell MA. 2019. Evolution of maternal lipid provisioning strategies in echinoids with non-feeding larvae: selection for high-quality juveniles. Mar Ecol Prog Ser. 616:95–106.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

