Stardust: improving spatial transcriptomics data analysis through space-aware modularity optimization-based clustering
- PMID: 35946989
- PMCID: PMC9364686
- DOI: 10.1093/gigascience/giac075
Stardust: improving spatial transcriptomics data analysis through space-aware modularity optimization-based clustering
Abstract
Background: Spatial transcriptomics (ST) combines stained tissue images with spatially resolved high-throughput RNA sequencing. The spatial transcriptomic analysis includes challenging tasks like clustering, where a partition among data points (spots) is defined by means of a similarity measure. Improving clustering results is a key factor as clustering affects subsequent downstream analysis. State-of-the-art approaches group data by taking into account transcriptional similarity and some by exploiting spatial information as well. However, it is not yet clear how much the spatial information combined with transcriptomics improves the clustering result.
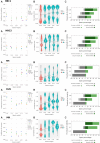
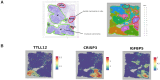
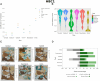
Results: We propose a new clustering method, Stardust, that easily exploits the combination of space and transcriptomic information in the clustering procedure through a manual or fully automatic tuning of algorithm parameters. Moreover, a parameter-free version of the method is also provided where the spatial contribution depends dynamically on the expression distances distribution in the space. We evaluated the proposed methods results by analyzing ST data sets available on the 10x Genomics website and comparing clustering performances with state-of-the-art approaches by measuring the spots' stability in the clusters and their biological coherence. Stability is defined by the tendency of each point to remain clustered with the same neighbors when perturbations are applied.
Conclusions: Stardust is an easy-to-use methodology allowing to define how much spatial information should influence clustering on different tissues and achieving more stable results than state-of-the-art approaches.
Keywords: clustering; spatial transcriptomics analysis; stability scores, parameters tuning, software comparison.
© The Author(s) 2022. Published by Oxford University Press GigaScience.
Figures


References
-
- Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–60. - PubMed
-
- Lewis SM, Asselin-Labat ML, Nguyen Q, et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat Methods. 2021;18(9):1–16. - PubMed
-
- Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353(6294):78–82. - PubMed
-
- Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes—next generation tools for tissue exploration. Bioessays. 2020;42(10):1900221. - PubMed
-
- Marx V. Method of the year: spatially resolved transcriptomics. Nat Methods. 2021;18(1):9–14. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

