Longitudinal expression profiling identifies a poor risk subset of patients with ABC-type diffuse large B-cell lymphoma
- PMID: 35947123
- PMCID: PMC9986713
- DOI: 10.1182/bloodadvances.2022007536
Longitudinal expression profiling identifies a poor risk subset of patients with ABC-type diffuse large B-cell lymphoma
Abstract
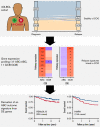
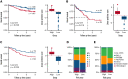
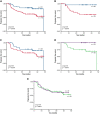
Despite the effectiveness of immuno-chemotherapy, 40% of patients with diffuse large B-cell lymphoma (DLBCL) experience relapse or refractory disease. Longitudinal studies have previously focused on the mutational landscape of relapse but fell short of providing a consistent relapse-specific genetic signature. In our study, we have focused attention on the changes in GEP accompanying DLBCL relapse using archival paired diagnostic/relapse specimens from 38 de novo patients with DLBCL. COO remained stable from diagnosis to relapse in 80% of patients, with only a single patient showing COO switching from activated B-cell-like (ABC) to germinal center B-cell-like (GCB). Analysis of the transcriptomic changes that occur following relapse suggest ABC and GCB relapses are mediated via different mechanisms. We developed a 30-gene discriminator for ABC-DLBCLs derived from relapse-associated genes that defined clinically distinct high- and low-risk subgroups in ABC-DLBCLs at diagnosis in datasets comprising both population-based and clinical trial cohorts. This signature also identified a population of <60-year-old patients with superior PFS and OS treated with ibrutinib-R-CHOP as part of the PHOENIX trial. Altogether this new signature adds to the existing toolkit of putative genetic predictors now available in DLBCL that can be readily assessed as part of prospective clinical trials.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: J.F. has provided consultancy and received funding from Epizyme. K.K. is an employee and shareholder of Roche. D.W.S. and L.M.R. have IP rights to the Lymph2Cx assay. D.W.S. has provided consultancy to AbbVie, AstraZeneca, Celgene, Janssen, and Incyte, and has received research funding from Janssen, NanoString Technology, and Roche/Genentech.
Figures


References
-
- Coiffier B, Lepage E, Brière J, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(4):235–242. - PubMed
-
- Ma Z, Niu J, Cao Y, et al. Clinical significance of ‘double-hit’ and ‘double-expression’ lymphomas. J Clin Pathol. 2019 jclinpath-2019-206199. - PubMed
-
- Liu Y, Barta SK. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am J Hematol. 2019;94(5):604–616. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

