Genomics reveal population structure, evolutionary history, and signatures of selection in the northern bottlenose whale, Hyperoodon ampullatus
- PMID: 35947506
- PMCID: PMC9804413
- DOI: 10.1111/mec.16643
Genomics reveal population structure, evolutionary history, and signatures of selection in the northern bottlenose whale, Hyperoodon ampullatus
Abstract
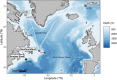
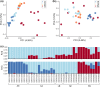
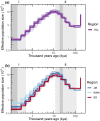
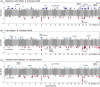
Information on wildlife population structure, demographic history, and adaptations are fundamental to understanding species evolution and informing conservation strategies. To study this ecological context for a cetacean of conservation concern, we conducted the first genomic assessment of the northern bottlenose whale, Hyperoodon ampullatus, using whole-genome resequencing data (n = 37) from five regions across the North Atlantic Ocean. We found a range-wide pattern of isolation-by-distance with a genetic subdivision distinguishing three subgroups: the Scotian Shelf, western North Atlantic, and Jan Mayen regions. Signals of elevated levels of inbreeding in the Endangered Scotian Shelf population indicate this population may be more vulnerable than the other two subgroups. In addition to signatures of inbreeding, evidence of local adaptation in the Scotian Shelf was detected across the genome. We found a long-term decline in effective population size for the species, which poses risks to their genetic diversity and may be exacerbated by the isolating effects of population subdivision. Protecting important habitat and migratory corridors should be prioritized to rebuild population sizes that were diminished by commercial whaling, strengthen gene flow, and ensure animals can move across regions in response to environmental changes.
Keywords: cetacean; conservation; genetic diversity; genomics; whale.
© 2022 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicting interests.
Figures

Similar articles
-
Evolutionary impacts differ between two exploited populations of northern bottlenose whale (Hyperoodon ampullatus).Ecol Evol. 2019 Nov 19;9(23):13567-13584. doi: 10.1002/ece3.5813. eCollection 2019 Dec. Ecol Evol. 2019. PMID: 31871667 Free PMC article.
-
The inference of gray whale (Eschrichtius robustus) historical population attributes from whole-genome sequences.BMC Evol Biol. 2018 Jun 7;18(1):87. doi: 10.1186/s12862-018-1204-3. BMC Evol Biol. 2018. PMID: 29879895 Free PMC article.
-
A comparison of genomic diversity and demographic history of the North Atlantic and Southwest Atlantic southern right whales.Mol Ecol. 2024 Oct;33(20):e17099. doi: 10.1111/mec.17099. Epub 2023 Aug 14. Mol Ecol. 2024. PMID: 37577945
-
Nuclear and mitochondrial markers reveal distinctiveness of a small population of bottlenose whales (Hyperoodon ampullatus) in the western North Atlantic.Mol Ecol. 2006 Oct;15(11):3115-29. doi: 10.1111/j.1365-294X.2006.03004.x. Mol Ecol. 2006. PMID: 16968258
-
Genomic Impact of Whaling in North Atlantic Fin Whales.Mol Biol Evol. 2022 May 3;39(5):msac094. doi: 10.1093/molbev/msac094. Mol Biol Evol. 2022. PMID: 35512360 Free PMC article.
Cited by
-
Ocean-Wide Conservation Genomics of Blue Whales Suggest New Northern Hemisphere Subspecies.Mol Ecol. 2025 Jan;34(2):e17619. doi: 10.1111/mec.17619. Epub 2024 Dec 17. Mol Ecol. 2025. PMID: 39688592 Free PMC article.
-
Evolutionary history and seascape genomics of Harbour porpoises (Phocoena phocoena) across environmental gradients in the North Atlantic and adjacent waters.Mol Ecol Resour. 2025 Jul;25(5):e13860. doi: 10.1111/1755-0998.13860. Epub 2023 Sep 8. Mol Ecol Resour. 2025. PMID: 37681405 Free PMC article.
-
The genome sequence of the Northern Bottlenose Whale, Hyperoodon ampullatus (Forster, 1770).Wellcome Open Res. 2024 Jul 26;9:410. doi: 10.12688/wellcomeopenres.22743.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39649623 Free PMC article.
-
Genetic architecture of long-distance migration and population genomics of the endangered Japanese eel.iScience. 2024 Jul 22;27(8):110563. doi: 10.1016/j.isci.2024.110563. eCollection 2024 Aug 16. iScience. 2024. PMID: 39165844 Free PMC article.
-
Aseasonal Migration of a Northern Bottlenose Whale Provides Support for the Skin Molt Migration Hypothesis.Ecol Evol. 2025 Jan 30;15(2):e70921. doi: 10.1002/ece3.70921. eCollection 2025 Feb. Ecol Evol. 2025. PMID: 39896783 Free PMC article.
References
-
- Baker, C. S. , Slade, R. W. , Bannister, J. L. , Abernethy, R. B. , Weinrich, M. T. , Lien, J. , Urban, J. , Corkeron, P. , Calmabokidis, K. , Vasquez, O. , & Palumbi, S. R. (1994). Hierarchical structure of mitochondrial DNA gene flow among humpback whales Megaptera novaeangliae, world‐wide. Molecular Ecology, 3(4), 313–327. - PubMed
-
- Bomba, L. , Nicolazzi, E. L. , Milanesi, M. , Negrini, R. , Mancini, G. , Biscarini, F. , Stella, A. , Valentini, A. , & Ajmone‐Marsan, P. (2015). Relative extended haplotype homozygosity signals across breeds reveal dairy and beef specific signatures of selection. Genetics Selection Evolution, 47, 25. 10.1186/s12711-015-0113-9 - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources

