An intermediate-effect size variant in UMOD confers risk for chronic kidney disease
- PMID: 35947615
- PMCID: PMC9388113
- DOI: 10.1073/pnas.2114734119
An intermediate-effect size variant in UMOD confers risk for chronic kidney disease
Abstract
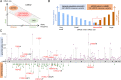
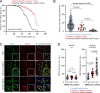
The kidney-specific gene UMOD encodes for uromodulin, the most abundant protein excreted in normal urine. Rare large-effect variants in UMOD cause autosomal dominant tubulointerstitial kidney disease (ADTKD), while common low-impact variants strongly associate with kidney function and the risk of chronic kidney disease (CKD) in the general population. It is unknown whether intermediate-effect variants in UMOD contribute to CKD. Here, candidate intermediate-effect UMOD variants were identified using large-population and ADTKD cohorts. Biological and phenotypical effects were investigated using cell models, in silico simulations, patient samples, and international databases and biobanks. Eight UMOD missense variants reported in ADTKD are present in the Genome Aggregation Database (gnomAD), with minor allele frequency (MAF) ranging from 10-5 to 10-3. Among them, the missense variant p.Thr62Pro is detected in ∼1/1,000 individuals of European ancestry, shows incomplete penetrance but a high genetic load in familial clusters of CKD, and is associated with kidney failure in the 100,000 Genomes Project (odds ratio [OR] = 3.99 [1.84 to 8.98]) and the UK Biobank (OR = 4.12 [1.32 to 12.85). Compared with canonical ADTKD mutations, the p.Thr62Pro carriers displayed reduced disease severity, with slower progression of CKD and an intermediate reduction of urinary uromodulin levels, in line with an intermediate trafficking defect in vitro and modest induction of endoplasmic reticulum (ER) stress. Identification of an intermediate-effect UMOD variant completes the spectrum of UMOD-associated kidney diseases and provides insights into the mechanisms of ADTKD and the genetic architecture of CKD.
Keywords: Autosomal Dominant Tubulointerstitial Kidney Disease; ER stress; genetic architecture; rare disease; uromodulin.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Uromodulin and CKD: insight into variant pathogenicity.Kidney Int. 2023 Feb;103(2):248-250. doi: 10.1016/j.kint.2022.11.018. Kidney Int. 2023. PMID: 36681451 No abstract available.
References
-
- Eckardt K.-U., et al. , Evolving importance of kidney disease: From subspecialty to global health burden. Lancet 382, 158–169 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

