The neuropeptide landscape of human prefrontal cortex
- PMID: 35947618
- PMCID: PMC9388106
- DOI: 10.1073/pnas.2123146119
The neuropeptide landscape of human prefrontal cortex
Abstract
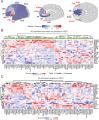
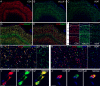
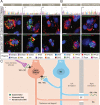
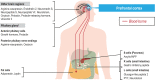
Human prefrontal cortex (hPFC) is a complex brain region involved in cognitive and emotional processes and several psychiatric disorders. Here, we present an overview of the distribution of the peptidergic systems in 17 subregions of hPFC and three reference cortices obtained by microdissection and based on RNA sequencing and RNAscope methods integrated with published single-cell transcriptomics data. We detected expression of 60 neuropeptides and 60 neuropeptide receptors in at least one of the hPFC subregions. The results reveal that the peptidergic landscape in PFC consists of closely located and functionally different subregions with unique peptide/transmitter-related profiles. Neuropeptide-rich PFC subregions were identified, encompassing regions from anterior cingulate cortex/orbitofrontal gyrus. Furthermore, marked differences in gene expression exist between different PFC regions (>5-fold; cocaine and amphetamine-regulated transcript peptide) as well as between PFC regions and reference regions, for example, for somatostatin and several receptors. We suggest that the present approach allows definition of, still hypothetical, microcircuits exemplified by glutamatergic neurons expressing a peptide cotransmitter either as an agonist (hypocretin/orexin) or antagonist (galanin). Specific neuropeptide receptors have been identified as possible targets for neuronal afferents and, interestingly, peripheral blood-borne peptide hormones (leptin, adiponectin, gastric inhibitory peptide, glucagon-like peptides, and peptide YY). Together with other recent publications, our results support the view that neuropeptide systems may play an important role in hPFC and underpin the concept that neuropeptide signaling helps stabilize circuit connectivity and fine-tune/modulate PFC functions executed during health and disease.
Keywords: RNA-seq; anterior cingulate cortex; classic neurotransmitter coexistence; in situ hybridization.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Fuster J., The Prefrontal Cortex (Academic Press, 2015).
-
- Dixon M. L., Thiruchselvam R., Todd R., Christoff K., Emotion and the prefrontal cortex: An integrative review. Psychol. Bull. 143, 1033–1081 (2017). - PubMed
-
- Ingvar D. H., Franzén G., Abnormalities of cerebral blood flow distribution in patients with chronic schizophrenia. Acta Psychiatr. Scand. 50, 425–462 (1974). - PubMed
-
- Berman K. F., Weinberger D. R., The prefrontal cortex in schizophrenia and other neuropsychiatric diseases: In vivo physiological correlates of cognitive deficits. Prog. Brain Res. 85, 521–536, discussion 536–537 (1990). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

