Single-cell multiomics sequencing reveals the reprogramming defects in embryos generated by round spermatid injection
- PMID: 35947654
- PMCID: PMC9365279
- DOI: 10.1126/sciadv.abm3976
Single-cell multiomics sequencing reveals the reprogramming defects in embryos generated by round spermatid injection
Abstract
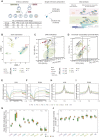
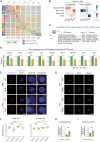
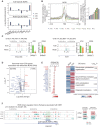
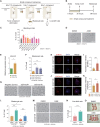
Round spermatid injection (ROSI) technique holds great promise for clinical treatment of a proportion of infertile men. However, the compromised developmental potential of ROSI embryos largely limits the clinical application, and the mechanisms are not fully understood. Here, we describe the transcriptome, chromatin accessibility, and DNA methylation landscapes of mouse ROSI embryos derived from early-stage round spermatids using a single-cell multiomics sequencing approach. By interrogating these data, we identify the reprogramming defects in ROSI embryos at the pronuclear stages, which are mainly associated with the misexpression of a cohort of minor zygotic genome activation genes. We screen a small compound, A366, that can significantly increase the developmental potential of ROSI embryos, in which A366 can partially overcome the reprogramming defects by amending the epigenetic and transcriptomic states. Collectively, our study uncovers the reprogramming defects in ROSI embryos for understanding the mechanisms underlying compromised developmental potential and offers an avenue for ROSI technique optimization.
Figures

Similar articles
-
How to improve the clinical outcome of round spermatid injection (ROSI) into the oocyte: Correction of epigenetic abnormalities.Reprod Med Biol. 2023 Feb 9;22(1):e12503. doi: 10.1002/rmb2.12503. eCollection 2023 Jan-Dec. Reprod Med Biol. 2023. PMID: 36789269 Free PMC article. Review.
-
Effects of the histone deacetylase inhibitor 'Scriptaid' on the developmental competence of mouse embryos generated through round spermatid injection.Hum Reprod. 2017 Jan;32(1):76-87. doi: 10.1093/humrep/dew290. Epub 2016 Nov 17. Hum Reprod. 2017. PMID: 27864358
-
Paternally inherited H3K27me3 affects chromatin accessibility in mouse embryos produced by round spermatid injection.Development. 2022 Sep 15;149(18):dev200696. doi: 10.1242/dev.200696. Epub 2022 Sep 15. Development. 2022. PMID: 35993297
-
Aberrant histone methylation in mouse early preimplantation embryos derived from round spermatid injection.Biochem Biophys Res Commun. 2023 Nov 5;680:119-126. doi: 10.1016/j.bbrc.2023.09.020. Epub 2023 Sep 12. Biochem Biophys Res Commun. 2023. PMID: 37738901
-
Clinical values and advances in round spermatid injection (ROSI).Reprod Biol. 2021 Sep;21(3):100530. doi: 10.1016/j.repbio.2021.100530. Epub 2021 Jun 23. Reprod Biol. 2021. PMID: 34171715 Review.
Cited by
-
Oocyte transcriptomes and follicular fluid proteomics of ovine atretic follicles reveal the underlying mechanisms of oocyte degeneration.BMC Genomics. 2025 Feb 1;26(1):97. doi: 10.1186/s12864-025-11291-9. BMC Genomics. 2025. PMID: 39893388 Free PMC article.
-
Single-Cell Transcriptome Analysis Reveals Development-Specific Networks at Distinct Synchronized Antral Follicle Sizes in Sheep Oocytes.Int J Mol Sci. 2024 Jan 11;25(2):910. doi: 10.3390/ijms25020910. Int J Mol Sci. 2024. PMID: 38255985 Free PMC article.
-
Single-cell multi-omics profiling of human preimplantation embryos identifies cytoskeletal defects during embryonic arrest.Nat Cell Biol. 2024 Feb;26(2):263-277. doi: 10.1038/s41556-023-01328-0. Epub 2024 Jan 18. Nat Cell Biol. 2024. PMID: 38238450
-
Single-cell multi-omics sequencing of human spermatogenesis reveals a DNA demethylation event associated with male meiotic recombination.Nat Cell Biol. 2023 Oct;25(10):1520-1534. doi: 10.1038/s41556-023-01232-7. Epub 2023 Sep 18. Nat Cell Biol. 2023. PMID: 37723297
-
How to improve the clinical outcome of round spermatid injection (ROSI) into the oocyte: Correction of epigenetic abnormalities.Reprod Med Biol. 2023 Feb 9;22(1):e12503. doi: 10.1002/rmb2.12503. eCollection 2023 Jan-Dec. Reprod Med Biol. 2023. PMID: 36789269 Free PMC article. Review.
References
-
- Willott G. M., Frequency of azoospermia. Forensic Sci. Int. 20, 9–10 (1982). - PubMed
-
- Gianaroli L., Selman H. A., Magli M. C., Colpi G., Fortini D., Ferraretti A. P., Birth of a healthy infant after conception with round spermatids isolated from cryopreserved testicular tissue. Fertil. Steril. 72, 539–541 (1999). - PubMed
-
- Mayer W., Niveleau A., Walter J., Fundele R., Haaf T., Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000). - PubMed
-
- Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J., Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 (2000). - PubMed
LinkOut - more resources
Full Text Sources

