Adenoviral-based vaccine promotes neoantigen-specific CD8+ T cell stemness and tumor rejection
- PMID: 35947675
- PMCID: PMC9844517
- DOI: 10.1126/scitranslmed.abo7604
Adenoviral-based vaccine promotes neoantigen-specific CD8+ T cell stemness and tumor rejection
Abstract
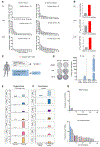
Upon chronic antigen exposure, CD8+ T cells become exhausted, acquiring a dysfunctional state correlated with the inability to control infection or tumor progression. In contrast, stem-like CD8+ T progenitors maintain the ability to promote and sustain effective immunity. Adenovirus (Ad)-vectored vaccines encoding tumor neoantigens have been shown to eradicate large tumors when combined with anti-programmed cell death protein 1 (αPD-1) in murine models; however, the mechanisms and translational potential have not yet been elucidated. Here, we show that gorilla Ad vaccine targeting tumor neoepitopes enhances responses to αPD-1 therapy by improving immunogenicity and antitumor efficacy. Single-cell RNA sequencing demonstrated that the combination of Ad vaccine and αPD-1 increased the number of murine polyfunctional neoantigen-specific CD8+ T cells over αPD-1 monotherapy, with an accumulation of Tcf1+ stem-like progenitors in draining lymph nodes and effector CD8+ T cells in tumors. Combined T cell receptor (TCR) sequencing analysis highlighted a broader spectrum of neoantigen-specific CD8+ T cells upon vaccination compared to αPD-1 monotherapy. The translational relevance of these data is supported by results obtained in the first 12 patients with metastatic deficient mismatch repair (dMMR) tumors vaccinated with an Ad vaccine encoding shared neoantigens. Expansion and diversification of TCRs were observed in post-treatment biopsies of patients with clinical response, as well as an increase in tumor-infiltrating T cells with an effector memory signature. These findings indicate a promising mechanism to overcome resistance to PD-1 blockade by promoting immunogenicity and broadening the spectrum and magnitude of neoantigen-specific T cells infiltrating tumors.
Figures






References
-
- Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, Forman J, Huang T, Li S, Zhang W, Xu Q, Cieri N, Clauser KR, Shukla SA, Neuberg D, Justesen S, MacBeath G, Carr SA, Fritsch EF, Hacohen N, Sade-Feldman M, Livak KJ, Boland GM, Ott PA, Keskin DB, Wu CJ, Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021). - PMC - PubMed
-
- He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, Xu L, Chen X, Hao Y, Wang P, Zhu C, Ou J, Liang H, Ni T, Zhang X, Zhou X, Deng K, Chen Y, Luo Y, Xu J, Qi H, Wu Y, Ye L, Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nature 537, 412–416 (2016). - PubMed
-
- Hudson WH, Gensheimer J, Hashimoto M, Wieland A, Valanparambil RM, Li P, Lin JX, Konieczny BT, Im SJ, Freeman GJ, Leonard WJ, Kissick HT, Ahmed R, Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity 51, 1043–1058.e4 (2019). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

