Rhamnosyltransferases involved in the biosynthesis of flavone rutinosides in Chrysanthemum species
- PMID: 35947689
- PMCID: PMC9706480
- DOI: 10.1093/plphys/kiac371
Rhamnosyltransferases involved in the biosynthesis of flavone rutinosides in Chrysanthemum species
Abstract
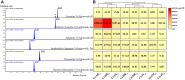
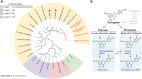
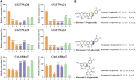
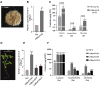
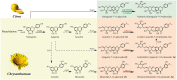
Linarin (acacetin-7-O-rutinoside), isorhoifolin (apigenin-7-O-rutinoside), and diosmin (diosmetin-7-O-rutinoside) are chemically and structurally similar flavone rutinoside (FR) compounds found in Chrysanthemum L. (Anthemideae, Asteraceae) plants. However, their biosynthetic pathways remain largely unknown. In this study, we cloned and compared FRs and genes encoding rhamnosyltransferases (RhaTs) among eight accessions of Chrysanthemum polyploids. We also biochemically characterized RhaTs of Chrysanthemum plants and Citrus (Citrus sinensis and Citrus maxima). RhaTs from these two genera are substrate-promiscuous enzymes catalyzing the rhamnosylation of flavones, flavanones, and flavonols. Substrate specificity analysis revealed that Chrysanthemum 1,6RhaTs preferred flavone glucosides (e.g. acacetin-7-O-glucoside), whereas Cs1,6RhaT preferred flavanone glucosides. The nonsynonymous substitutions of RhaTs found in some cytotypes of diploids resulted in the loss of catalytic function. Phylogenetic analysis and specialized pathways responsible for the biosynthesis of major flavonoids in Chrysanthemum and Citrus revealed that rhamnosylation activity might share a common evolutionary origin. Overexpression of RhaT in hairy roots resulted in 13-, 2-, and 5-fold increases in linarin, isorhoifolin, and diosmin contents, respectively, indicating that RhaT is mainly involved in the biosynthesis of linarin. Our findings not only suggest that the substrate promiscuity of RhaTs contributes to the diversity of FRs in Chrysanthemum species but also shed light on the evolution of flavone and flavanone rutinosides in distant taxa.
© The Author(s) 2022. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures



Similar articles
-
Collinearity analysis and characterization of rhamnosyltransferases from Chrysanthemum morifolium, Mikania micrantha and Stevia rebaudiana.Planta. 2025 Jun 13;262(2):24. doi: 10.1007/s00425-025-04742-w. Planta. 2025. PMID: 40512260
-
The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: evolution of branch-forming rhamnosyltransferases under domestication.Plant J. 2013 Jan;73(1):166-78. doi: 10.1111/tpj.12030. Epub 2012 Nov 8. Plant J. 2013. PMID: 22989156
-
Substrate preference of citrus naringenin rhamnosyltransferases and their application to flavonoid glycoside production in fission yeast.Appl Microbiol Biotechnol. 2016 Jan;100(2):687-96. doi: 10.1007/s00253-015-6982-6. Epub 2015 Oct 3. Appl Microbiol Biotechnol. 2016. PMID: 26433966
-
Pharmacology of Diosmin, a Citrus Flavone Glycoside: An Updated Review.Eur J Drug Metab Pharmacokinet. 2022 Jan;47(1):1-18. doi: 10.1007/s13318-021-00731-y. Epub 2021 Oct 23. Eur J Drug Metab Pharmacokinet. 2022. PMID: 34687440 Review.
-
Update on uses and properties of citrus flavonoids: new findings in anticancer, cardiovascular, and anti-inflammatory activity.J Agric Food Chem. 2008 Aug 13;56(15):6185-205. doi: 10.1021/jf8006568. Epub 2008 Jul 2. J Agric Food Chem. 2008. PMID: 18593176 Review.
Cited by
-
Rhamnosyltransferases in Chrysanthemum: Just a spoonful of sugar helps the flavonoid-based medicines abound.Plant Physiol. 2022 Nov 28;190(4):2061-2063. doi: 10.1093/plphys/kiac457. Plant Physiol. 2022. PMID: 36149328 Free PMC article. No abstract available.
-
Collinearity analysis and characterization of rhamnosyltransferases from Chrysanthemum morifolium, Mikania micrantha and Stevia rebaudiana.Planta. 2025 Jun 13;262(2):24. doi: 10.1007/s00425-025-04742-w. Planta. 2025. PMID: 40512260
-
Co-Expression Network Analysis of the Transcriptome Identified Hub Genes and Pathways Responding to Saline-Alkaline Stress in Sorghum bicolor L.Int J Mol Sci. 2023 Nov 27;24(23):16831. doi: 10.3390/ijms242316831. Int J Mol Sci. 2023. PMID: 38069156 Free PMC article.
-
Tailored biosynthesis of diosmin through reconstitution of the flavonoid pathway in Nicotiana benthamiana.Front Plant Sci. 2024 Oct 18;15:1464877. doi: 10.3389/fpls.2024.1464877. eCollection 2024. Front Plant Sci. 2024. PMID: 39494057 Free PMC article.
-
Comparative Analysis of the Chemical Constituents of Chrysanthemum morifolium with Different Drying Processes Integrating LC/GC-MS-Based, Non-Targeted Metabolomics.Metabolites. 2024 Sep 2;14(9):481. doi: 10.3390/metabo14090481. Metabolites. 2024. PMID: 39330488 Free PMC article.
References
-
- Albohy A, Zahran EM, Abdelmohsen UR, Salem MA, Al-Warhi T, Al-Sanea MM, Abelyan N, Khalil HE, Desoukey SY, Fouad MA, et al. (2020) Multitarget in silico studies of Ocimum menthiifolium, family Lamiaceae against SARS-CoV-2 supported by molecular dynamics simulation. J Biomol Struct Dyn 40: 4062–4072 - PMC - PubMed
-
- Ballester AR, Lafuente MT (2017) LED blue light-induced changes in phenolics and ethylene in citrus fruit: implication in elicited resistance against Penicillium digitatum infection. Food Chem 218: 575–583 - PubMed
-
- Bansal P, Paul P, Mudgal J, Nayak PG, Pannakal ST, Priyadarsini KI, Unnikrishnan MK (2012) Antidiabetic, antihyperlipidemic and antioxidant effects of the flavonoid rich fraction of Pilea microphylla (L.) in high fat diet/streptozotocin-induced diabetes in mice. Exp Toxicol Pathol 64: 651–658 - PubMed
-
- Baris O, Karadayi M, Yanmis D, Guvenalp Z, Bal T, Gulluce M (2011) Isolation of 3 flavonoids from Mentha longifolia (L.) Hudson subsp. longifolia and determination of their genotoxic potentials by using the E. coli WP2 test system. J Food Sci 76: T212–T217 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

