Discovery of compounds that reactivate p53 mutants in vitro and in vivo
- PMID: 35948006
- PMCID: PMC9481737
- DOI: 10.1016/j.chembiol.2022.07.003
Discovery of compounds that reactivate p53 mutants in vitro and in vivo
Abstract
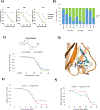
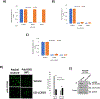
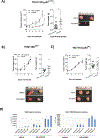
The tumor suppressor p53 is the most frequently mutated protein in human cancer. The majority of these mutations are missense mutations in the DNA binding domain of p53. Restoring p53 tumor suppressor function could have a major impact on the therapy for a wide range of cancers. Here we report a virtual screening approach that identified several small molecules with p53 reactivation activities. The UCI-LC0023 compound series was studied in detail and was shown to bind p53, induce a conformational change in mutant p53, restore the ability of p53 hotspot mutants to associate with chromatin, reestablish sequence-specific DNA binding of a p53 mutant in a reconstituted in vitro system, induce p53-dependent transcription programs, and prevent progression of tumors carrying mutant p53, but not p53null or p53WT alleles. Our study demonstrates feasibility of a computation-guided approach to identify small molecule corrector drugs for p53 hotspot mutations.
Keywords: cryptic pocket; ensemble based virtual screening; molecular dynamics simulations; mutant p53; p53 reactivation; small molecule p53 corrector drugs.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests Several authors of this manuscript are listed as inventors on the patent listed below. The patent describes compounds reported in this manuscript. Amaro, R. E., Baronio, R., Demir, O., Kaiser, P., Lathrop, R. H., Salehi-Amiri, S.-F., Wassman, C. Small molecules to enhance p53 activity. US patent US20160193214 A1. Approved March 2017.
Figures



References
-
- Beraza N, and Trautwein C (2007). Restoration of p53 function: a new therapeutic strategy to induce tumor regression? Hepatology 45, 1578–1579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

