YfeX - A New Platform for Carbene Transferase Development with High Intrinsic Reactivity
- PMID: 35948517
- PMCID: PMC9691539
- DOI: 10.1002/chem.202201474
YfeX - A New Platform for Carbene Transferase Development with High Intrinsic Reactivity
Abstract
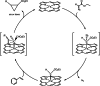
Carbene transfer biocatalysis has evolved from basic science to an area with vast potential for the development of new industrial processes. In this study, we show that YfeX, naturally a peroxidase, has great potential for the development of new carbene transferases, due to its high intrinsic reactivity, especially for the N-H insertion reaction of aromatic and aliphatic primary and secondary amines. YfeX shows high stability against organic solvents (methanol and DMSO), greatly improving turnover of hydrophobic substrates. Interestingly, in styrene cyclopropanation, WT YfeX naturally shows high enantioselectivity, generating the trans product with 87 % selectivity for the (R,R) enantiomer. WT YfeX also catalyzes the Si-H insertion efficiently. Steric effects in the active site were further explored using the R232A variant. Quantum Mechanics/Molecular Mechanics (QM/MM) calculations reveal details on the mechanism of Si-H insertion. YfeX, and potentially other peroxidases, are exciting new targets for the development of improved carbene transferases.
Keywords: N−H insertion; QM/MM calculations; biocatalysis; carbene transfer; cyclopropanation.
© 2022 The Authors. Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Chemoselective Cyclopropanation over Carbene Y-H Insertion Catalyzed by an Engineered Carbene Transferase.J Org Chem. 2018 Jul 20;83(14):7480-7490. doi: 10.1021/acs.joc.8b00946. Epub 2018 Jul 6. J Org Chem. 2018. PMID: 29905476 Free PMC article.
-
Reversing the Enantioselectivity of Enzymatic Carbene N-H Insertion Through Mechanism-Guided Protein Engineering.Angew Chem Int Ed Engl. 2023 Aug 28;62(35):e202303879. doi: 10.1002/anie.202303879. Epub 2023 Jul 20. Angew Chem Int Ed Engl. 2023. PMID: 37260412
-
Chemoselective N-H insertion catalyzed by a de novo carbene transferase.Biotechnol Appl Biochem. 2020 Jul;67(4):527-535. doi: 10.1002/bab.1924. Epub 2020 May 22. Biotechnol Appl Biochem. 2020. PMID: 32277840
-
Navigating the Unnatural Reaction Space: Directed Evolution of Heme Proteins for Selective Carbene and Nitrene Transfer.Acc Chem Res. 2021 Mar 2;54(5):1209-1225. doi: 10.1021/acs.accounts.0c00591. Epub 2021 Jan 25. Acc Chem Res. 2021. PMID: 33491448 Free PMC article. Review.
-
Carbene Transfer Reactions Catalysed by Dyes of the Metalloporphyrin Group.Molecules. 2018 Mar 29;23(4):792. doi: 10.3390/molecules23040792. Molecules. 2018. PMID: 29596367 Free PMC article. Review.
Cited by
-
Mechanistic investigation of sustainable heme-inspired biocatalytic synthesis of cyclopropanes for challenging substrates.Commun Chem. 2024 Nov 29;7(1):279. doi: 10.1038/s42004-024-01371-4. Commun Chem. 2024. PMID: 39613908 Free PMC article.
-
A comprehensive mechanistic investigation of sustainable carbene N-H insertion catalyzed by engineered His-ligated heme proteins.Catal Sci Technol. 2025 Jan 13;15(6):1802-1813. doi: 10.1039/d4cy00999a. eCollection 2025 Mar 17. Catal Sci Technol. 2025. PMID: 39882070 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

