Absolute binding free energy calculations improve enrichment of actives in virtual compound screening
- PMID: 35948614
- PMCID: PMC9365818
- DOI: 10.1038/s41598-022-17480-w
Absolute binding free energy calculations improve enrichment of actives in virtual compound screening
Abstract
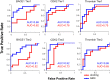
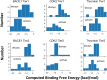
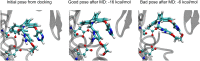
We determined the effectiveness of absolute binding free energy (ABFE) calculations to refine the selection of active compounds in virtual compound screening, a setting where the more commonly used relative binding free energy approach is not readily applicable. To do this, we conducted baseline docking calculations of structurally diverse compounds in the DUD-E database for three targets, BACE1, CDK2 and thrombin, followed by ABFE calculations for compounds with high docking scores. The docking calculations alone achieved solid enrichment of active compounds over decoys. Encouragingly, the ABFE calculations then improved on this baseline. Analysis of the results emphasizes the importance of establishing high quality ligand poses as starting points for ABFE calculations, a nontrivial goal when processing a library of diverse compounds without informative co-crystal structures. Overall, our results suggest that ABFE calculations can play a valuable role in the drug discovery process.
© 2022. The Author(s).
Conflict of interest statement
MKG has an equity interest in and is a cofounder and scientic advisor of VeraChem LLC. MF and GH have no competing interests.
Figures