Genetically-biased fertilization in APOBEC1 complementation factor (A1cf) mutant mice
- PMID: 35948620
- PMCID: PMC9365768
- DOI: 10.1038/s41598-022-17948-9
Genetically-biased fertilization in APOBEC1 complementation factor (A1cf) mutant mice
Abstract
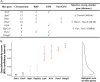
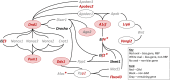
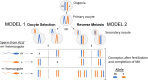
Meiosis, recombination, and gametogenesis normally ensure that gametes combine randomly. But in exceptional cases, fertilization depends on the genetics of gametes from both females and males. A key question is whether their non-random union results from factors intrinsic to oocytes and sperm, or from their interactions with conditions in the reproductive tracts. To address this question, we used in vitro fertilization (IVF) with a mutant and wild-type allele of the A1cf (APOBEC1 complementation factor) gene in mice that are otherwise genetically identical. We observed strong distortion in favor of mutant heterozygotes showing that bias depends on the genetics of oocyte and sperm, and that any environmental input is modest. To search for the potential mechanism of the 'biased fertilization', we analyzed the existing transcriptome data and demonstrated that localization of A1cf transcripts and its candidate mRNA targets is restricted to the spermatids in which they originate, and that these transcripts are enriched for functions related to meiosis, fertilization, RNA stability, translation, and mitochondria. We propose that failure to sequester mRNA targets in A1cf mutant heterozygotes leads to functional differences among spermatids, thereby providing an opportunity for selection among haploid gametes. The study adds to the understanding of the gamete interaction at fertilization. Discovery that bias is evident with IVF provides a new venue for future explorations of preference among genetically distinct gametes at fertilization for A1cf and other genes that display significant departure of Mendelian inheritance.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bateson W, Mendel G. Mendel’s Principles of Heredity. Cambridge University Press; 1902.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

