MEK1/2 inhibition rescues neurodegeneration by TFEB-mediated activation of autophagic lysosomal function in a model of Alzheimer's Disease
- PMID: 35948663
- PMCID: PMC9734062
- DOI: 10.1038/s41380-022-01713-5
MEK1/2 inhibition rescues neurodegeneration by TFEB-mediated activation of autophagic lysosomal function in a model of Alzheimer's Disease
Abstract
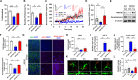
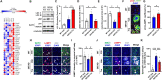
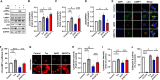
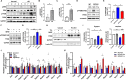
Alzheimer's Disease (AD) is a progressive neurodegenerative disorder, which is characterized by cognitive deficit due to synaptic loss and neuronal death. Extracellular amyloid β plaques are one of the pathological hallmarks of AD. The autophagic lysosomal pathway is the essential mechanism to maintain cellular homeostasis by driving clearance of protein aggregates and is dysfunctional in AD. Here, we showed that inhibiting MEK/ERK signaling using a clinically available MEK1/2 inhibitor, trametinib (GSK1120212, SNR1611), induces the protection of neurons through autophagic lysosomal activation mediated by transcription factor EB (TFEB) in a model of AD. Orally administered trametinib recovered impaired neural structures, cognitive functions, and hippocampal long-term potentiation (LTP) in 5XFAD mice. Trametinib also reduced Aβ deposition via induction of autophagic lysosomal activation. RNA-sequencing analysis revealed upregulation of autophagic lysosomal genes by trametinib administration. In addition, trametinib inhibited TFEB phosphorylation at Ser142 and promoted its nuclear translocation, which in turn induced autophagic lysosomal related genes, indicating that trametinib activates the autophagic lysosomal process through TFEB activation. From these observations, we concluded that MEK inhibition provides neuronal protection from the Aβ burden by increasing autophagic lysosomal activity. Thus, MEK inhibition may be an effective therapeutic strategy for AD.
© 2022. The Author(s).
Conflict of interest statement
MYK and SH are inventors of PCT/KR2017/013444 and its family patent applications regarding the use of trametinib for neurodegenerative disease. MYK, TJH, and SH are stockholders of Genuv Inc. The authors declare no competing interest.
Figures

References
-
- Sala Frigerio C, De Strooper B. Alzheimer’s Disease Mechanisms And Emerging Roads To Novel Therapeutics. Annu Rev Neurosci. 2016;39:57–79. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

