Clonal evolution in primary breast cancers under sequential epirubicin and docetaxel monotherapy
- PMID: 35948919
- PMCID: PMC9367103
- DOI: 10.1186/s13073-022-01090-2
Clonal evolution in primary breast cancers under sequential epirubicin and docetaxel monotherapy
Abstract
Background: Subclonal evolution during primary breast cancer treatment is largely unexplored. We aimed to assess the dynamic changes in subclonal composition of treatment-naïve breast cancers during neoadjuvant chemotherapy.
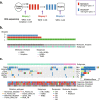
Methods: We performed whole exome sequencing of tumor biopsies collected before, at therapy switch, and after treatment with sequential epirubicin and docetaxel monotherapy in 51 out of 109 patients with primary breast cancer, who were included in a prospectively registered, neoadjuvant single-arm phase II trial.
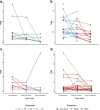
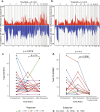
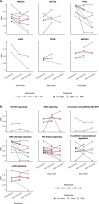
Results: There was a profound and differential redistribution of subclones during epirubicin and docetaxel treatment, regardless of therapy response. While truncal mutations and main subclones persisted, smaller subclones frequently appeared or disappeared. Reassessment of raw data, beyond formal mutation calling, indicated that the majority of subclones seemingly appearing during treatment were in fact present in pretreatment breast cancers, below conventional detection limits. Likewise, subclones which seemingly disappeared were still present, below detection limits, in most cases where tumor tissue remained. Tumor mutational burden (TMB) dropped during neoadjuvant therapy, and copy number analysis demonstrated specific genomic regions to be systematically lost or gained for each of the two chemotherapeutics.
Conclusions: Sequential epirubicin and docetaxel monotherapy caused profound redistribution of smaller subclones in primary breast cancer, while early truncal mutations and major subclones generally persisted through treatment.
Trial registration: ClinicalTrials.gov, NCT00496795 , registered on July 4, 2007.
Keywords: Breast cancer; Chemoresistance; Clonal evolution; Neoadjuvant chemotherapy.
© 2022. The Author(s).
Conflict of interest statement
JG received honoraria for consulting, advisory role, speaker’s bureau, and/or research funding from AstraZeneca, BMS, Daiichi Sankyo, Eli Lilly, MSD, Novartis, Pfizer, Pierre Fabre, and Roche, all outside the submitted work. PEL received honoraria for consulting, advisory role, speaker’s bureau, and/or research funding from AbbVie, Akademikonferens, AstraZeneca, Farmaceuticos Rovi, Illumina, Laboratorios, Novartis, Pfizer, Pierre-Fabre, and Roche, all outside the submitted work. SK received honoraria for consulting, advisory role, speaker’s bureau, and/or research funding from AstraZeneca, Illumina, Pfizer, and Pierre Fabre, all outside the submitted work. HPE received honoraria for consulting, advisory role, speaker’s bureau, and/or research funding from Amgen, Aptitude Health, AstraZeneca, BMS, Daiichi Sankyo, Eli Lilly, Hai Interaktiv, MSD, Novartis, Pfizer, Pierre Fabre, and Roche, all outside the submitted work. The remaining authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

