Multicolor plate reader fluorescence calibration
- PMID: 35949424
- PMCID: PMC9357555
- DOI: 10.1093/synbio/ysac010
Multicolor plate reader fluorescence calibration
Abstract
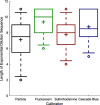
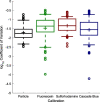
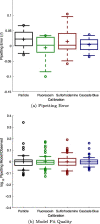
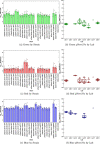
Plate readers are commonly used to measure cell growth and fluorescence, yet the utility and reproducibility of plate reader data is limited by the fact that it is typically reported in arbitrary or relative units. We have previously established a robust serial dilution protocol for calibration of plate reader measurements of absorbance to estimated bacterial cell count and for green fluorescence from proteins expressed in bacterial cells to molecules of equivalent fluorescein. We now extend these protocols to calibration of red fluorescence to the sulforhodamine-101 fluorescent dye and blue fluorescence to Cascade Blue. Evaluating calibration efficacy via an interlaboratory study, we find that these calibrants do indeed provide comparable precision to the prior calibrants and that they enable effective cross-laboratory comparison of measurements of red and blue fluorescence from proteins expressed in bacterial cells.
Keywords: calibration; cell count; fluorescence; units.
© The Author(s) 2022. Published by Oxford University Press.
Figures


References
-
- Auld D.S., Coassin P.A., Coussens N.P., Hensley P., Klumpp-Thomas C., Michael S., Sitta Sittampalam G., Joseph Trask O., Wagner B.K. and Weidner J.R.. et al. (2020) Microplate selection and recommended practices in high-throughput screening and quantitative biology. Assay Guidance Manual [Internet].
-
- Beal J. (2017) Biochemical complexity drives log-normal variation in genetic expression. Eng. Biol., 1, 55–60.doi: 10.1049/enb.2017.0004. - DOI
-
- Beal J., Baldwin G.S., Farny N.G., Gershater M., Haddock-Angelli T., Buckley-Taylor R., Dwijayanti A., Kiga D., Lizarazo M. and Marken J.. et al.. (2021) Comparative analysis of three studies measuring fluorescence from engineered bacterial genetic constructs. PloS One, 16, e0252263.doi: 10.1371/journal.pone.0252263. - DOI - PMC - PubMed
-
- Beal J., Haddock-Angelli T., Baldwin G., Gershater M., Dwijayanti A., Storch M., Kim M., Lizarazo M., Rettberg R. and the iGEM Interlab Study Contributors (2018) Quantification of bacterial fluorescence using independent calibrants. PloS One, 13, e0199432.doi: 10.1371/journal.pone.0199432. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
