Identification of potential key molecules and signaling pathways for psoriasis based on weighted gene co-expression network analysis
- PMID: 35949853
- PMCID: PMC9254198
- DOI: 10.12998/wjcc.v10.i18.5965
Identification of potential key molecules and signaling pathways for psoriasis based on weighted gene co-expression network analysis
Abstract
Background: Psoriasis is a chronic inflammatory skin disease, the pathogenesis of which is more complicated and often requires long-term treatment. In particular, moderate to severe psoriasis usually requires systemic treatment. Psoriasis is also associated with many diseases, such as cardiometabolic diseases, malignant tumors, infections, and mood disorders. Psoriasis can appear at any age, and lead to a substantial burden for individuals and society. At present, psoriasis is still a treatable, but incurable, disease. Previous studies have found that microRNAs (miRNAs) play an important regulatory role in the progression of various diseases. Currently, miRNAs studies in psoriasis and dermatology are relatively new. Therefore, the identification of key miRNAs in psoriasis is helpful to elucidate the molecular mechanism of psoriasis.
Aim: To identify key molecular markers and signaling pathways to provide potential basis for the treatment and management of psoriasis.
Methods: The miRNA and mRNA data were obtained from the Gene Expression Omnibus database. Then, differentially expressed mRNAs (DEmRNAs) and differentially expressed miRNAs (DEmiRNAs) were screened out by limma R package. Subsequently, DEmRNAs were analyzed for Gene Ontology and Kyoto Encyclopedia of Genes and Genomics functional enrichment. The "WGCNA" R package was used to analyze the co-expression network of all miRNAs. In addition, we constructed miRNA-mRNA regulatory networks based on identified hub miRNAs. Finally, in vitro validation was performed. All experimental procedures were approved by the ethics committee of Chinese PLA General Hospital (S2021-012-01).
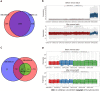
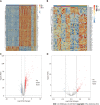
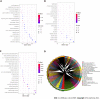
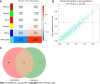
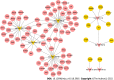
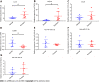
Results: A total of 639 DEmRNAs and 84 DEmiRNAs were identified. DEmRNAs screening criteria were adjusted P (adj. P) value < 0.01 and |logFoldChange| (|logFC|) > 1. DEmiRNAs screening criteria were adj. P value < 0.01 and |logFC| > 1.5. KEGG functional analysis demonstrated that DEmRNAs were significantly enriched in immune-related biological functions, for example, toll-like receptor signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway. In weighted gene co-expression network analysis, turquoise module was the hub module. Moreover, 10 hub miRNAs were identified. Among these 10 hub miRNAs, only 8 hub miRNAs predicted the corresponding target mRNAs. 97 negatively regulated miRNA-mRNA pairs were involved in the miRNA-mRNA regulatory network, for example, hsa-miR-21-5p-claudin 8 (CLDN8), hsa-miR-30a-3p-interleukin-1B (IL-1B), and hsa-miR-181a-5p/hsa-miR-30c-2-3p-C-X-C motif chemokine ligand 9 (CXCL9). Real-time polymerase chain reaction results showed that IL-1B and CXCL9 were up-regulated and CLDN8 was down-regulated in psoriasis with statistically significant differences.
Conclusion: The identification of potential key molecular markers and signaling pathways provides potential research directions for further understanding the molecular mechanisms of psoriasis. This may also provide new research ideas for the prevention and treatment of psoriasis in the future.
Keywords: Functional enrichment; MicroRNA-mRNA regulatory network; MicroRNAs; Psoriasis; Weighted gene co-expression network analysis.
©The Author(s) 2022. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Figures


Similar articles
-
Construction of circRNA-Based ceRNA Network to Reveal the Role of circRNAs in the Progression and Prognosis of Hepatocellular Carcinoma.Front Genet. 2021 Feb 26;12:626764. doi: 10.3389/fgene.2021.626764. eCollection 2021. Front Genet. 2021. PMID: 33719338 Free PMC article.
-
Uncovering of Key Pathways and miRNAs for Intracranial Aneurysm Based on Weighted Gene Co-Expression Network Analysis.Eur Neurol. 2022;85(3):212-223. doi: 10.1159/000521390. Epub 2022 Jan 14. Eur Neurol. 2022. PMID: 35034029
-
Identification of a Potentially Functional microRNA-mRNA Regulatory Network in Lung Adenocarcinoma Using a Bioinformatics Analysis.Front Cell Dev Biol. 2021 Feb 18;9:641840. doi: 10.3389/fcell.2021.641840. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33681226 Free PMC article.
-
Network pharmacology-based identification of miRNA expression of Astragalus membranaceus in the treatment of diabetic nephropathy.Medicine (Baltimore). 2022 Feb 4;101(5):e28747. doi: 10.1097/MD.0000000000028747. Medicine (Baltimore). 2022. PMID: 35119030 Free PMC article.
-
Meta-analysis of transcriptomics data identifies potential biomarkers and their associated regulatory networks in gallbladder cancer.Gastroenterol Hepatol Bed Bench. 2022;15(4):311-325. doi: 10.22037/ghfbb.v15i4.2292. Gastroenterol Hepatol Bed Bench. 2022. PMID: 36762219 Free PMC article. Review.
Cited by
-
Molecular Morbidity Score-Can MicroRNAs Assess the Burden of Disease?Int J Mol Sci. 2024 Jul 24;25(15):8042. doi: 10.3390/ijms25158042. Int J Mol Sci. 2024. PMID: 39125612 Free PMC article. Review.
-
Design and Evaluation of Tretinoin Fatty Acid Vesicles for the Topical Treatment of Psoriasis.Molecules. 2023 Nov 30;28(23):7868. doi: 10.3390/molecules28237868. Molecules. 2023. PMID: 38067597 Free PMC article.
-
Monocytes subsets altered distribution and dysregulated plasma hsa-miR-21-5p and hsa-miR-155-5p in HCV-linked liver cirrhosis progression to hepatocellular carcinoma.J Cancer Res Clin Oncol. 2023 Nov;149(17):15349-15364. doi: 10.1007/s00432-023-05313-w. Epub 2023 Aug 28. J Cancer Res Clin Oncol. 2023. PMID: 37639012 Free PMC article.
References
-
- López-Estebaranz JL, Sánchez-Carazo JL, Sulleiro S. Effect of a family history of psoriasis and age on comorbidities and quality of life in patients with moderate to severe psoriasis: Results from the ARIZONA study. J Dermatol. 2016;43:395–401. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

