Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6
- PMID: 35950458
- PMCID: PMC9385563
- DOI: 10.14348/molcells.2022.0040
Structural Insights into Porphyrin Recognition by the Human ATP-Binding Cassette Transporter ABCB6
Abstract
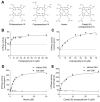
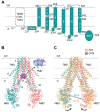
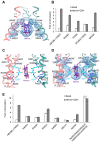
Human ABCB6 is an ATP-binding cassette transporter that regulates heme biosynthesis by translocating various porphyrins from the cytoplasm into the mitochondria. Here we report the cryo-electron microscopy (cryo-EM) structures of human ABCB6 with its substrates, coproporphyrin III (CPIII) and hemin, at 3.5 and 3.7 Å resolution, respectively. Metalfree porphyrin CPIII binds to ABCB6 within the central cavity, where its propionic acids form hydrogen bonds with the highly conserved Y550. The resulting structure has an overall fold similar to the inward-facing apo structure, but the two nucleotide-binding domains (NBDs) are slightly closer to each other. In contrast, when ABCB6 binds a metal-centered porphyrin hemin in complex with two glutathione molecules (1 hemin: 2 glutathione), the two NBDs end up much closer together, aligning them to bind and hydrolyze ATP more efficiently. In our structures, a glycine-rich and highly flexible "bulge" loop on TM helix 7 undergoes significant conformational changes associated with substrate binding. Our findings suggest that ABCB6 utilizes at least two distinct mechanisms to fine-tune substrate specificity and transport efficiency.
Keywords: ABCB6; ATP-binding cassette transporter; cryoelectron microscopy; glutathione; porphyrin.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures



Similar articles
-
Cryo-EM structure of cadmium-bound human ABCB6.Commun Biol. 2024 May 31;7(1):672. doi: 10.1038/s42003-024-06377-1. Commun Biol. 2024. PMID: 38822018 Free PMC article.
-
Cryo-electron microscopy structure of human ABCB6 transporter.Protein Sci. 2020 Dec;29(12):2363-2374. doi: 10.1002/pro.3960. Epub 2020 Oct 15. Protein Sci. 2020. PMID: 33007128 Free PMC article.
-
W546 stacking disruption traps the human porphyrin transporter ABCB6 in an outward-facing transient state.Commun Biol. 2023 Sep 21;6(1):960. doi: 10.1038/s42003-023-05339-3. Commun Biol. 2023. PMID: 37735522 Free PMC article.
-
The role of ABCG2 and ABCB6 in porphyrin metabolism and cell survival.Curr Pharm Biotechnol. 2011 Apr;12(4):647-55. doi: 10.2174/138920111795163995. Curr Pharm Biotechnol. 2011. PMID: 21118089 Review.
-
ABCB6, an ABC Transporter Impacting Drug Response and Disease.AAPS J. 2017 Nov 30;20(1):8. doi: 10.1208/s12248-017-0165-6. AAPS J. 2017. PMID: 29192381 Free PMC article. Review.
Cited by
-
Capture of endogenous lipids in peptidiscs and effect on protein stability and activity.iScience. 2024 Mar 1;27(4):109382. doi: 10.1016/j.isci.2024.109382. eCollection 2024 Apr 19. iScience. 2024. PMID: 38577106 Free PMC article.
-
Cryo-EM structure of cadmium-bound human ABCB6.Commun Biol. 2024 May 31;7(1):672. doi: 10.1038/s42003-024-06377-1. Commun Biol. 2024. PMID: 38822018 Free PMC article.
-
The role of ATP-binding Cassette subfamily B member 6 in the inner ear.Nat Commun. 2024 Nov 18;15(1):9885. doi: 10.1038/s41467-024-53663-x. Nat Commun. 2024. PMID: 39557842 Free PMC article.
-
ATP-Binding Cassette and Solute Carrier Transporters: Understanding Their Mechanisms and Drug Modulation Through Structural and Modeling Approaches.Pharmaceuticals (Basel). 2024 Nov 27;17(12):1602. doi: 10.3390/ph17121602. Pharmaceuticals (Basel). 2024. PMID: 39770445 Free PMC article. Review.
-
Emerging Role of ABC Transporters in Glia Cells in Health and Diseases of the Central Nervous System.Cells. 2024 Apr 24;13(9):740. doi: 10.3390/cells13090740. Cells. 2024. PMID: 38727275 Free PMC article. Review.
References
-
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

