The HPV Induced Cancer Resource (THInCR): a Suite of Tools for Investigating HPV-Dependent Human Carcinogenesis
- PMID: 35950764
- PMCID: PMC9429961
- DOI: 10.1128/msphere.00317-22
The HPV Induced Cancer Resource (THInCR): a Suite of Tools for Investigating HPV-Dependent Human Carcinogenesis
Abstract
Human papillomaviruses (HPVs) are highly infectious and cause the most common sexually transmitted viral infections. They induce hyperproliferation of squamous epithelial tissue, often forming warts. Virally encoded proteins reprogram gene expression and cell growth to create an optimal environment for viral replication. In addition to their normal roles in infection, functional alterations induced by viral proteins establish conditions that frequently contribute to human carcinogenesis. In fact, ~5% of human cancers are caused by HPVs, with virtually all cervical squamous cell carcinomas (CESC) and an increasing number of head and neck squamous cell carcinomas (HNSC) attributed to HPV infection. The Cancer Genome Atlas (TCGA) molecularly characterized thousands of primary human cancer samples in many cancer types, including CESC and HNSC, and created a comprehensive atlas of genomic, epigenomic, and transcriptomic data. This publicly available genome-wide information provides an unprecedented opportunity to expand the knowledge of the role that HPV plays in human carcinogenesis. While many tools exist to mine these data, few, if any, focus on the comparison of HPV-positive cancers with their HPV-negative counterparts or adjacent normal control tissue. We have constructed a suite of web-based tools, The HPV Induced Cancer Resource (THInCR), to utilize TCGA data for research related to HPV-induced CESC and HNSC. These tools allow investigators to gain greater biological and medical insights by exploring the impacts of HPV on cellular gene expression (mRNA and microRNA), altered gene methylation, and associations with patient survival and immune landscape features. These tools are accessible at https://thincr.ca/. IMPORTANCE The suite of analytical tools of THInCR provides the opportunity to investigate the roles that candidate target genes identified in cell lines or other model systems contribute to in actual HPV-dependent human cancers and is based on large-scale TCGA data sets. Expression of target genes, including both mRNA and microRNA, can be correlated with HPV gene expression, epigenetic changes in DNA methylation, patient survival, and numerous immune features, like leukocyte infiltration, interferon gamma response, T cell response, etc. Data from these analyses may immediately provide evidence to validate in vitro observations, reveal insights into mechanisms of virus-mediated alterations in cell growth, behavior, gene expression, and innate and adaptive immunity and may help hypothesis generation for further investigations.
Keywords: DNA methylation; HPV; TCGA; analysis resource; cancer; cervical cancer; correlation; database; gene expression; head and neck cancer; human papillomavirus; methylation; oncogene; survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
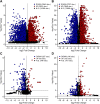
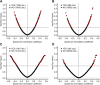
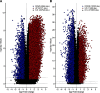
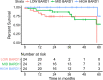
Similar articles
-
Combined Transcriptome and Proteome Analysis of Immortalized Human Keratinocytes Expressing Human Papillomavirus 16 (HPV16) Oncogenes Reveals Novel Key Factors and Networks in HPV-Induced Carcinogenesis.mSphere. 2019 Mar 27;4(2):e00129-19. doi: 10.1128/mSphere.00129-19. mSphere. 2019. PMID: 30918060 Free PMC article.
-
Bioinformatics analysis of immune characteristics in tumors with alternative carcinogenesis pathways induced by human papillomaviruses.Virol J. 2023 Dec 4;20(1):287. doi: 10.1186/s12985-023-02241-6. Virol J. 2023. PMID: 38049810 Free PMC article.
-
DNA Methylation and mRNA Expression of OX40 (TNFRSF4) and GITR (TNFRSF18, AITR) in Head and Neck Squamous Cell Carcinoma Correlates With HPV Status, Mutational Load, an Interferon-γ Signature, Signatures of Immune Infiltrates, and Survival.J Immunother. 2022 May 1;45(4):194-206. doi: 10.1097/CJI.0000000000000407. J Immunother. 2022. PMID: 34908008
-
HPV Integration in Head and Neck Squamous Cell Carcinomas: Cause and Consequence.Recent Results Cancer Res. 2017;206:57-72. doi: 10.1007/978-3-319-43580-0_4. Recent Results Cancer Res. 2017. PMID: 27699529 Review.
-
APOBEC-mediated genomic alterations link immunity and viral infection during human papillomavirus-driven cervical carcinogenesis.Biosci Trends. 2017 Sep 12;11(4):383-388. doi: 10.5582/bst.2017.01103. Epub 2017 Jul 17. Biosci Trends. 2017. PMID: 28717061 Review.
Cited by
-
Repurposing therapy of ibrexafungerp vulvovaginal candidiasis drugs as cancer therapeutics.Front Pharmacol. 2024 Jun 27;15:1428755. doi: 10.3389/fphar.2024.1428755. eCollection 2024. Front Pharmacol. 2024. PMID: 38994207 Free PMC article. No abstract available.
-
The role of the SOX2 gene in cervical cancer: focus on ferroptosis and construction of a predictive model.J Cancer Res Clin Oncol. 2024 Nov 23;150(12):509. doi: 10.1007/s00432-024-05973-2. J Cancer Res Clin Oncol. 2024. PMID: 39580372 Free PMC article.
-
Human papilloma virus infection drives unique metabolic and immune profiles in head and neck and cervical cancers: implications for targeted therapies and prognostic markers.Discov Oncol. 2025 May 6;16(1):676. doi: 10.1007/s12672-025-02384-8. Discov Oncol. 2025. PMID: 40327200 Free PMC article.
-
The EBV Gastric Cancer Resource (EBV-GCR): A Suite of Tools for Investigating EBV-Associated Human Gastric Carcinogenesis.Viruses. 2023 Mar 27;15(4):853. doi: 10.3390/v15040853. Viruses. 2023. PMID: 37112833 Free PMC article.
-
The viral etiology of EBV-associated gastric cancers contributes to their unique pathology, clinical outcomes, treatment responses and immune landscape.Front Immunol. 2024 Mar 26;15:1358511. doi: 10.3389/fimmu.2024.1358511. eCollection 2024. Front Immunol. 2024. PMID: 38596668 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

