Systems spatiotemporal dynamics of traumatic brain injury at single-cell resolution reveals humanin as a therapeutic target
- PMID: 35951114
- PMCID: PMC9372016
- DOI: 10.1007/s00018-022-04495-9
Systems spatiotemporal dynamics of traumatic brain injury at single-cell resolution reveals humanin as a therapeutic target
Abstract
Background: The etiology of mild traumatic brain injury (mTBI) remains elusive due to the tissue and cellular heterogeneity of the affected brain regions that underlie cognitive impairments and subsequent neurological disorders. This complexity is further exacerbated by disrupted circuits within and between cell populations across brain regions and the periphery, which occur at different timescales and in spatial domains.
Methods: We profiled three tissues (hippocampus, frontal cortex, and blood leukocytes) at the acute (24-h) and subacute (7-day) phases of mTBI at single-cell resolution.
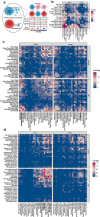
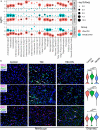
Results: We demonstrated that the coordinated gene expression patterns across cell types were disrupted and re-organized by TBI at different timescales with distinct regional and cellular patterns. Gene expression-based network modeling implied astrocytes as a key regulator of the cell-cell coordination following mTBI in both hippocampus and frontal cortex across timepoints, and mt-Rnr2, which encodes the mitochondrial peptide humanin, as a potential target for intervention based on its broad regional and dynamic dysregulation following mTBI. Treatment of a murine mTBI model with humanin reversed cognitive impairment caused by mTBI through the restoration of metabolic pathways within astrocytes.
Conclusions: Our results offer a systems-level understanding of the dynamic and spatial regulation of gene programs by mTBI and pinpoint key target genes, pathways, and cell circuits that are amenable to therapeutics.
Keywords: Astrocytes; Humanin; Mt-Rnr2; Single-cell RNA sequencing; TBI; Traumatic brain injury.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Yamaki T, Imahori Y, Ohmori Y, Yoshino E, Hohri T, Ebisu T, Ueda S. Cerebral hemodynamics and metabolism of severe diffuse brain injury measured by PET. J Nucl Med. 1996;37:1166–1170. - PubMed
-
- Bergsneider M, Hovda DA, Lee SM, Kelly DF, McArthur DL, Vespa PM, Lee JH, Huang SC, Martin NA, Phelps ME, Becker DP. Dissociation of cerebral glucose metabolism and level of consciousness during the period of metabolic depression following human traumatic brain injury. J Neurotrauma. 2000;17:389–401. doi: 10.1089/neu.2000.17.389. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

