Replication dependent and independent mechanisms of GAA repeat instability
- PMID: 35952488
- PMCID: PMC9675320
- DOI: 10.1016/j.dnarep.2022.103385
Replication dependent and independent mechanisms of GAA repeat instability
Abstract
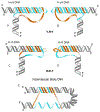
Trinucleotide repeat instability is a driver of human disease. Large expansions of (GAA)n repeats in the first intron of the FXN gene are the cause Friedreich's ataxia (FRDA), a progressive degenerative disorder which cannot yet be prevented or treated. (GAA)n repeat instability arises during both replication-dependent processes, such as cell division and intergenerational transmission, as well as in terminally differentiated somatic tissues. Here, we provide a brief historical overview on the discovery of (GAA)n repeat expansions and their association to FRDA, followed by recent advances in the identification of triplex H-DNA formation and replication fork stalling. The main body of this review focuses on the last decade of progress in understanding the mechanism of (GAA)n repeat instability during DNA replication and/or DNA repair. We propose that the discovery of additional mechanisms of (GAA)n repeat instability can be achieved via both comparative approaches to other repeat expansion diseases and genome-wide association studies. Finally, we discuss the advances towards FRDA prevention or amelioration that specifically target (GAA)n repeat expansions.
Keywords: DNA triplex; Friedreich’s ataxia; GAA repeat instability; H-DNA; Replication fork stalling; Trinucleotide repeats.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare that there are no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

