Ultra-deep sequencing validates safety of CRISPR/Cas9 genome editing in human hematopoietic stem and progenitor cells
- PMID: 35953477
- PMCID: PMC9372057
- DOI: 10.1038/s41467-022-32233-z
Ultra-deep sequencing validates safety of CRISPR/Cas9 genome editing in human hematopoietic stem and progenitor cells
Abstract
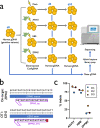
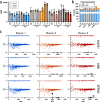
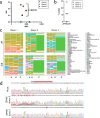
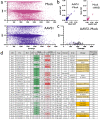
As CRISPR-based therapies enter the clinic, evaluation of safety remains a critical and active area of study. Here, we employ a clinical next generation sequencing (NGS) workflow to achieve high sequencing depth and detect ultra-low frequency variants across exons of genes associated with cancer, all exons, and genome wide. In three separate primary human hematopoietic stem and progenitor cell (HSPC) donors assessed in technical triplicates, we electroporated high-fidelity Cas9 protein targeted to three loci (AAVS1, HBB, and ZFPM2) and harvested genomic DNA at days 4 and 10. Our results demonstrate that clinically relevant delivery of high-fidelity Cas9 to primary HSPCs and ex vivo culture up to 10 days does not introduce or enrich for tumorigenic variants and that even a single SNP in a gRNA spacer sequence is sufficient to eliminate Cas9 off-target activity in primary, repair-competent human HSPCs.
© 2022. The Author(s).
Conflict of interest statement
The authors of this study declare the following competing interests: M.H.P. is on the Board of Directors of Graphite Bio. M.H.P. serves on the SAB of Allogene Tx and is an advisor to Versant Ventures. M.H.P., M.K.C., and V.V.B. hold equity in Graphite Bio. V.V.B. serves on the Board of Directors and SAB of Umoja Biopharma, the Board of Directors of ArsenalBio, and is a board observer at Synthego. M.H.P. holds equity in CRISPR Tx. E.J., M.W., F.C., I.K., A. G., and T.T. are employees of Illumina, Inc. The remaining authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

