TET enzymes regulate skeletal development through increasing chromatin accessibility of RUNX2 target genes
- PMID: 35953487
- PMCID: PMC9372040
- DOI: 10.1038/s41467-022-32138-x
TET enzymes regulate skeletal development through increasing chromatin accessibility of RUNX2 target genes
Abstract
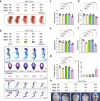
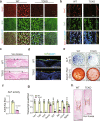
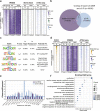
The Ten-eleven translocation (TET) family of dioxygenases mediate cytosine demethylation by catalyzing the oxidation of 5-methylcytosine (5mC). TET-mediated DNA demethylation controls the proper differentiation of embryonic stem cells and TET members display functional redundancy during early gastrulation. However, it is unclear if TET proteins have functional significance in mammalian skeletal development. Here, we report that Tet genes deficiency in mesoderm mesenchymal stem cells results in severe defects of bone development. The existence of any single Tet gene allele can support early bone formation, suggesting a functional redundancy of TET proteins. Integrative analyses of RNA-seq, Whole Genome Bisulfite Sequencing (WGBS), 5hmC-Seal and Assay for Transposase-Accessible Chromatin (ATAC-seq) demonstrate that TET-mediated demethylation increases the chromatin accessibility of target genes by RUNX2 and facilities RUNX2-regulated transcription. In addition, TET proteins interact with RUNX2 through their catalytic domain to regulate cytosine methylation around RUNX2 binding region. The catalytic domain is indispensable for TET enzymes to regulate RUNX2 transcription activity on its target genes and to regulate bone development. These results demonstrate that TET enzymes function to regulate RUNX2 activity and maintain skeletal homeostasis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling.Nature. 2016 Oct 27;538(7626):528-532. doi: 10.1038/nature20095. Epub 2016 Oct 19. Nature. 2016. PMID: 27760115
-
Tet family of 5-methylcytosine dioxygenases in mammalian development.J Hum Genet. 2013 Jul;58(7):421-7. doi: 10.1038/jhg.2013.63. Epub 2013 May 30. J Hum Genet. 2013. PMID: 23719188 Free PMC article. Review.
-
Mechanisms that regulate the activities of TET proteins.Cell Mol Life Sci. 2022 Jun 15;79(7):363. doi: 10.1007/s00018-022-04396-x. Cell Mol Life Sci. 2022. PMID: 35705880 Free PMC article. Review.
-
Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA.Science. 2011 Sep 2;333(6047):1303-7. doi: 10.1126/science.1210944. Epub 2011 Aug 4. Science. 2011. PMID: 21817016 Free PMC article.
-
Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.Nature. 2014 Feb 20;506(7488):391-5. doi: 10.1038/nature12905. Epub 2013 Dec 25. Nature. 2014. PMID: 24390346 Free PMC article.
Cited by
-
DNMT3L inhibits hepatocellular carcinoma progression through DNA methylation of CDO1: insights from big data to basic research.J Transl Med. 2024 Feb 2;22(1):128. doi: 10.1186/s12967-024-04939-9. J Transl Med. 2024. PMID: 38308276 Free PMC article.
-
Effect of DNA methylation on the osteogenic differentiation of mesenchymal stem cells: concise review.Front Genet. 2024 Jul 2;15:1429844. doi: 10.3389/fgene.2024.1429844. eCollection 2024. Front Genet. 2024. PMID: 39015772 Free PMC article. Review.
-
Cell signaling and transcriptional regulation of osteoblast lineage commitment, differentiation, bone formation, and homeostasis.Cell Discov. 2024 Jul 2;10(1):71. doi: 10.1038/s41421-024-00689-6. Cell Discov. 2024. PMID: 38956429 Free PMC article. Review.
-
Cellular stress and epigenetic regulation in adult stem cells.Life Sci Alliance. 2024 Sep 30;7(12):e202302083. doi: 10.26508/lsa.202302083. Print 2024 Dec. Life Sci Alliance. 2024. PMID: 39348938 Free PMC article. Review.
-
Cell identity and 5-hydroxymethylcytosine.Epigenetics Chromatin. 2025 Jun 19;18(1):36. doi: 10.1186/s13072-025-00601-w. Epigenetics Chromatin. 2025. PMID: 40537872 Free PMC article. Review.
References
-
- Chakraborty, R. et al. Histone acetyltransferases p300 and CBP coordinate distinct chromatin remodeling programs in vascular smooth muscle plasticity. Circulation145, 1720–1737 (2022). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

