Innate Immune Response and Inflammasome Activation During SARS-CoV-2 Infection
- PMID: 35953688
- PMCID: PMC9371632
- DOI: 10.1007/s10753-022-01651-y
Innate Immune Response and Inflammasome Activation During SARS-CoV-2 Infection
Abstract
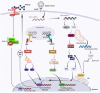
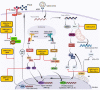
The novel coronavirus SARS-CoV-2, responsible for the COVID-19 outbreak, has become a pandemic threatening millions of lives worldwide. Recently, several vaccine candidates and drugs have shown promising effects in preventing or treating COVID-19, but due to the development of mutant strains through rapid viral evolution, urgent investigations are warranted in order to develop preventive measures and further improve current vaccine candidates. Positive-sense-single-stranded RNA viruses comprise many (re)emerging human pathogens that pose a public health problem. Our innate immune system and, in particular, the interferon response form an important first line of defense against these viruses. Flexibility in the genome aids the virus to develop multiple strategies to evade the innate immune response and efficiently promotes their replication and infective capacity. This review will focus on the innate immune response to SARS-CoV-2 infection and the virus' evasion of the innate immune system by escaping recognition or inhibiting the production of an antiviral state. Since interferons have been implicated in inflammatory diseases and immunopathology along with their protective role in infection, antagonizing the immune response may have an ambiguous effect on the clinical outcome of the viral disease. This pathology is characterized by intense, rapid stimulation of the innate immune response that triggers activation of the Nod-like receptor family, pyrin-domain-containing 3 (NLRP3) inflammasome pathway, and release of its products including the pro-inflammatory cytokines IL-6, IL-18, and IL-1β. This predictive view may aid in designing an immune intervention or preventive vaccine for COVID-19 in the near future.
Keywords: COVID-19; Inflammasome; Innate immune response; NLRP3; SARS-CoV2; Vaccine..
© 2022. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

