Griottes: a generalist tool for network generation from segmented tissue images
- PMID: 35953853
- PMCID: PMC9367069
- DOI: 10.1186/s12915-022-01376-2
Griottes: a generalist tool for network generation from segmented tissue images
Abstract
Background: Microscopy techniques and image segmentation algorithms have improved dramatically this decade, leading to an ever increasing amount of biological images and a greater reliance on imaging to investigate biological questions. This has created a need for methods to extract the relevant information on the behaviors of cells and their interactions, while reducing the amount of computing power required to organize this information.
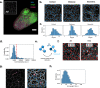
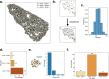
Results: This task can be performed by using a network representation in which the cells and their properties are encoded in the nodes, while the neighborhood interactions are encoded by the links. Here, we introduce Griottes, an open-source tool to build the "network twin" of 2D and 3D tissues from segmented microscopy images. We show how the library can provide a wide range of biologically relevant metrics on individual cells and their neighborhoods, with the objective of providing multi-scale biological insights. The library's capacities are demonstrated on different image and data types.
Conclusions: This library is provided as an open-source tool that can be integrated into common image analysis workflows to increase their capacities.
Keywords: Graphs; Image analysis; Napari; Python; Single-cell imaging; Spatial analysis; Tissue imaging.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

