Neuroinflammatory Gene Expression Analysis Reveals Pathways of Interest as Potential Targets to Improve the Recording Performance of Intracortical Microelectrodes
- PMID: 35954192
- PMCID: PMC9367362
- DOI: 10.3390/cells11152348
Neuroinflammatory Gene Expression Analysis Reveals Pathways of Interest as Potential Targets to Improve the Recording Performance of Intracortical Microelectrodes
Abstract
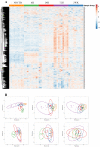
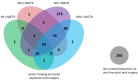
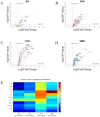
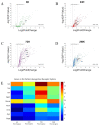
Intracortical microelectrodes are a critical component of brain-machine interface (BMI) systems. The recording performance of intracortical microelectrodes used for both basic neuroscience research and clinical applications of BMIs decreases over time, limiting the utility of the devices. The neuroinflammatory response to the microelectrode has been identified as a significant contributing factor to its performance. Traditionally, pathological assessment has been limited to a dozen or so known neuroinflammatory proteins, and only a few groups have begun to explore changes in gene expression following microelectrode implantation. Our initial characterization of gene expression profiles of the neuroinflammatory response to mice implanted with non-functional intracortical probes revealed many upregulated genes that could inform future therapeutic targets. Emphasis was placed on the most significant gene expression changes and genes involved in multiple innate immune sets, including Cd14, C3, Itgam, and Irak4. In previous studies, inhibition of Cluster of Differentiation 14 (Cd14) improved microelectrode performance for up to two weeks after electrode implantation, suggesting CD14 can be explored as a potential therapeutic target. However, all measures of improvements in signal quality and electrode performance lost statistical significance after two weeks. Therefore, the current study investigated the expression of genes in the neuroinflammatory pathway at the tissue-microelectrode interface in Cd14-/- mice to understand better how Cd14 inhibition was connected to temporary improvements in recording quality over the initial 2-weeks post-surgery, allowing for the identification of potential co-therapeutic targets that may work synergistically with or after CD14 inhibition to improve microelectrode performance.
Keywords: Toll-like receptors; cluster of differentiation 14; complement; cytokine; extracellular matrix; inflammation; microelectrode.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



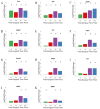
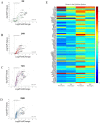


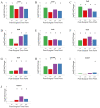
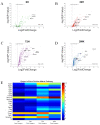
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

