Identification, Quantification, and Characterization of HIV-1 Reservoirs in the Human Brain
- PMID: 35954221
- PMCID: PMC9367788
- DOI: 10.3390/cells11152379
Identification, Quantification, and Characterization of HIV-1 Reservoirs in the Human Brain
Abstract
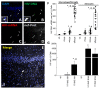
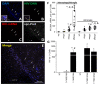
The major barrier to cure HIV infection is the early generation and extended survival of HIV reservoirs in the circulation and tissues. Currently, the techniques used to detect and quantify HIV reservoirs are mostly based on blood-based assays; however, it has become evident that viral reservoirs remain in tissues. Our study describes a novel multi-component imaging method (HIV DNA, mRNA, and viral proteins in the same assay) to identify, quantify, and characterize viral reservoirs in tissues and blood products obtained from HIV-infected individuals even when systemic replication is undetectable. In the human brains of HIV-infected individuals under ART, we identified that microglia/macrophages and a small population of astrocytes are the main cells with integrated HIV DNA. Only half of the cells with integrated HIV DNA expressed viral mRNA, and one-third expressed viral proteins. Surprisingly, we identified residual HIV-p24, gp120, nef, vpr, and tat protein expression and accumulation in uninfected cells around HIV-infected cells suggesting local synthesis, secretion, and bystander uptake. In conclusion, our data show that ART reduces the size of the brain's HIV reservoirs; however, local/chronic viral protein secretion still occurs, indicating that the brain is still a major anatomical target to cure HIV infection.
Keywords: HIV-1; NeuroHIV; anti-retroviral; cure; dementia; reservoirs; survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
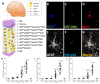
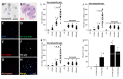
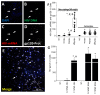
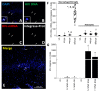

Similar articles
-
Astrocytes are HIV reservoirs in the brain: A cell type with poor HIV infectivity and replication but efficient cell-to-cell viral transfer.J Neurochem. 2021 Jul;158(2):429-443. doi: 10.1111/jnc.15336. Epub 2021 Mar 22. J Neurochem. 2021. PMID: 33655498 Free PMC article.
-
Ultrasensitive HIV-1 p24 Assay Detects Single Infected Cells and Differences in Reservoir Induction by Latency Reversal Agents.J Virol. 2017 Feb 28;91(6):e02296-16. doi: 10.1128/JVI.02296-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077644 Free PMC article.
-
Identification, Localization, and Quantification of HIV Reservoirs Using Microscopy.Curr Protoc Cell Biol. 2019 Mar;82(1):e64. doi: 10.1002/cpcb.64. Epub 2018 Sep 28. Curr Protoc Cell Biol. 2019. PMID: 30265439 Free PMC article.
-
Modulation of apoptosis and viral latency - an axis to be well understood for successful cure of human immunodeficiency virus.J Gen Virol. 2016 Apr;97(4):813-824. doi: 10.1099/jgv.0.000402. Epub 2016 Jan 13. J Gen Virol. 2016. PMID: 26764023 Review.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
Cited by
-
A Rationale and Approach to the Development of Specific Treatments for HIV Associated Neurocognitive Impairment.Microorganisms. 2022 Nov 12;10(11):2244. doi: 10.3390/microorganisms10112244. Microorganisms. 2022. PMID: 36422314 Free PMC article. Review.
-
Antiretroviral drugs from multiple classes induce loss of excitatory synapses between hippocampal neurons in culture.Front Pharmacol. 2024 Mar 12;15:1369757. doi: 10.3389/fphar.2024.1369757. eCollection 2024. Front Pharmacol. 2024. PMID: 38533258 Free PMC article.
-
Epigenetic modulation of myeloid cell functions in HIV and SARS-CoV-2 infection.Mol Biol Rep. 2024 Feb 24;51(1):342. doi: 10.1007/s11033-024-09266-2. Mol Biol Rep. 2024. PMID: 38400997 Free PMC article. Review.
-
PROTAC-mediated degradation of HIV-1 Nef efficiently restores cell-surface CD4 and MHC-I expression and blocks HIV-1 replication.Cell Chem Biol. 2024 Apr 18;31(4):658-668.e14. doi: 10.1016/j.chembiol.2024.02.004. Epub 2024 Mar 19. Cell Chem Biol. 2024. PMID: 38508197 Free PMC article.
-
Increasing Neuroinflammation Relates to Increasing Neurodegeneration in People with HIV.Viruses. 2023 Aug 30;15(9):1835. doi: 10.3390/v15091835. Viruses. 2023. PMID: 37766242 Free PMC article.
References
-
- De Scheerder M.A., Vrancken B., Dellicour S., Schlub T., Lee E., Shao W., Rutsaert S., Verhofstede C., Kerre T., Malfait T., et al. HIV Rebound Is Predominantly Fueled by Genetically Identical Viral Expansions from Diverse Reservoirs. Cell Host Microbe. 2019;26:347–358.e347. doi: 10.1016/j.chom.2019.08.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

