Preliminary Discovery of Small Molecule Inhibitors of Epidermal Growth Factor Receptor (EGFR) That Bind to the Extracellular Domain
- PMID: 35954311
- PMCID: PMC9367601
- DOI: 10.3390/cancers14153647
Preliminary Discovery of Small Molecule Inhibitors of Epidermal Growth Factor Receptor (EGFR) That Bind to the Extracellular Domain
Abstract
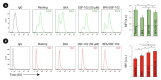
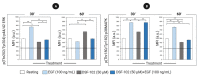
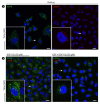
The Epidermal Growth Factor Receptor (EGFR) is a transmembrane glycoprotein belonging to the protein kinase superfamily. It is composed of an extracellular domain, a transmembrane anchoring region and a cytoplasmic region endowed with tyrosine kinase activity. Genetic mutations of EGFR kinase cause higher activity thereby stimulating downstream signaling pathways that, in turn, impact transcription and cell cycle progression. Due to the involvement of mutant EGFR in tumors and inflammatory diseases, in the past decade, several EGFR inhibitory strategies have been extensively studied, either targeting the extracellular domain (through monoclonal antibodies) or the intracellular kinase domain (through ATP-mimic small molecules). Monoclonal antibodies impair the binding to growth factor, the receptor dimerization, and its activation, whereas small molecules block the intracellular catalytic activity. Herein, we describe the development of a novel small molecule, called DSF-102, that interacts with the extracellular domain of EGFR. When tested in vitro in KRAS mutant A549 cells, it impairs EGFR activity by exerting (i) dose-dependent toxicity effects; (ii) a negative regulation of ERK, MAPK p38 and AKT; and (iii) a modulation of the intracellular trafficking and lysosomal degradation of EGFR. Interestingly, DSF-102 exerts its EGFR inhibitory activity without showing interaction with the intracellular kinase domain. Taken together, these findings suggest that DSF-102 is a promising hit compound for the development of a novel class of anti-EGFR compounds, i.e., small molecules able to interact with the extracellular domain of EGFR and useful for overcoming the KRAS-driven resistance to TKI treatment.
Keywords: EGFR-ECD; inhibition; isatin; trafficking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The ErbB/HER family of protein-tyrosine kinases and cancer.Pharmacol Res. 2014 Jan;79:34-74. doi: 10.1016/j.phrs.2013.11.002. Epub 2013 Nov 20. Pharmacol Res. 2014. PMID: 24269963 Review.
-
Small molecule inhibitors targeting the EGFR/ErbB family of protein-tyrosine kinases in human cancers.Pharmacol Res. 2019 Jan;139:395-411. doi: 10.1016/j.phrs.2018.11.014. Epub 2018 Nov 27. Pharmacol Res. 2019. PMID: 30500458 Review.
-
Resistance to small molecule inhibitors of epidermal growth factor receptor in malignant gliomas.Cancer Res. 2003 Nov 1;63(21):7443-50. Cancer Res. 2003. PMID: 14612544
-
Growth hormone alters epidermal growth factor receptor binding affinity via activation of extracellular signal-regulated kinases in 3T3-F442A cells.Endocrinology. 2004 Jul;145(7):3297-306. doi: 10.1210/en.2003-1658. Epub 2004 Apr 7. Endocrinology. 2004. PMID: 15070853
-
Cetuximab: an epidermal growth factor receptor chemeric human-murine monoclonal antibody.Drugs Today (Barc). 2005 Feb;41(2):107-27. doi: 10.1358/dot.2005.41.2.882662. Drugs Today (Barc). 2005. PMID: 15821783 Review.
Cited by
-
Recent Advancements, Limitations, and Future Perspectives of the use of Personalized Medicine in Treatment of Colon Cancer.Technol Cancer Res Treat. 2023 Jan-Dec;22:15330338231178403. doi: 10.1177/15330338231178403. Technol Cancer Res Treat. 2023. PMID: 37248615 Free PMC article. Review.
-
Landscape and clinical implications of EGFR exon 20 insertions in non-small cell lung cancer patients.Clin Transl Oncol. 2025 Apr 4. doi: 10.1007/s12094-025-03899-w. Online ahead of print. Clin Transl Oncol. 2025. PMID: 40186089 Review.
-
EGFRp4 Peptides: A Novel Strategy for Epidermal Growth Factor Receptor (EGFR) Inhibitor.ACS Omega. 2025 Jun 16;10(25):26848-26856. doi: 10.1021/acsomega.5c01684. eCollection 2025 Jul 1. ACS Omega. 2025. PMID: 40620947 Free PMC article.
-
Isorhamnetin Regulates Programmed Death Ligand-1 Expression by Suppressing the EGFR-STAT3 Signaling Pathway in Canine Mammary Tumors.Int J Mol Sci. 2024 Jan 4;25(1):670. doi: 10.3390/ijms25010670. Int J Mol Sci. 2024. PMID: 38203840 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

