Comparative Multicentric Evaluation of Inter-Observer Variability in Manual and Automatic Segmentation of Neuroblastic Tumors in Magnetic Resonance Images
- PMID: 35954314
- PMCID: PMC9367307
- DOI: 10.3390/cancers14153648
Comparative Multicentric Evaluation of Inter-Observer Variability in Manual and Automatic Segmentation of Neuroblastic Tumors in Magnetic Resonance Images
Abstract
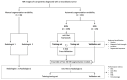
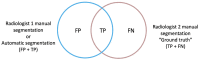
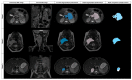
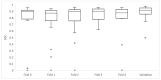
Tumor segmentation is one of the key steps in imaging processing. The goals of this study were to assess the inter-observer variability in manual segmentation of neuroblastic tumors and to analyze whether the state-of-the-art deep learning architecture nnU-Net can provide a robust solution to detect and segment tumors on MR images. A retrospective multicenter study of 132 patients with neuroblastic tumors was performed. Dice Similarity Coefficient (DSC) and Area Under the Receiver Operating Characteristic Curve (AUC ROC) were used to compare segmentation sets. Two more metrics were elaborated to understand the direction of the errors: the modified version of False Positive (FPRm) and False Negative (FNR) rates. Two radiologists manually segmented 46 tumors and a comparative study was performed. nnU-Net was trained-tuned with 106 cases divided into five balanced folds to perform cross-validation. The five resulting models were used as an ensemble solution to measure training (n = 106) and validation (n = 26) performance, independently. The time needed by the model to automatically segment 20 cases was compared to the time required for manual segmentation. The median DSC for manual segmentation sets was 0.969 (±0.032 IQR). The median DSC for the automatic tool was 0.965 (±0.018 IQR). The automatic segmentation model achieved a better performance regarding the FPRm. MR images segmentation variability is similar between radiologists and nnU-Net. Time leverage when using the automatic model with posterior visual validation and manual adjustment corresponds to 92.8%.
Keywords: automatic segmentation; deep learning; inter-observer variability; manual segmentation; neuroblastic tumors; tumor segmentation.
Conflict of interest statement
All authors certify that there is no actual or potential conflict of interest in relation to this article.
Figures

References
-
- Cohn S.L., Pearson A.D.J., London W.B., Monclair T., Ambros P.F., Brodeur G.M., Faldum A., Hero B., Iehara T., Machin D., et al. The International Neuroblastoma Risk Group (INRG) Classification System: An INRG Task Force Report. J. Clin. Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. - DOI - PMC - PubMed
-
- Brisse H.J., McCarville M.B., Granata C., Krug K.B., Wootton-Gorges S.L., Kanegawa K., Giammarile F., Schmidt M., Shulkin B., Matthay K.K., et al. Guidelines for Imaging and Staging of Neuroblastic Tumors: Consensus Report from the International Neuroblastoma Risk Group Project. Radiology. 2011;261:243–257. doi: 10.1148/radiol.11101352. - DOI - PubMed
-
- Martí-Bonmatí L., Alberich-Bayarri Á., Ladenstein R., Blanquer I., Segrelles J.D., Cerdá-Alberich L., Gkontra P., Hero B., Garcia-Aznar J.M., Keim D., et al. PRIMAGE project: Predictive in silico multiscale analytics to support childhood cancer personalised evaluation empowered by imaging biomarkers. Eur. Radiol. Exp. 2020;4:22. doi: 10.1186/s41747-020-00150-9. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

