Live Cell Detection of Poly(ADP-Ribose) for Use in Genetic and Genotoxic Compound Screens
- PMID: 35954352
- PMCID: PMC9367489
- DOI: 10.3390/cancers14153676
Live Cell Detection of Poly(ADP-Ribose) for Use in Genetic and Genotoxic Compound Screens
Abstract
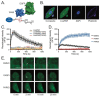
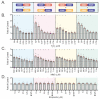
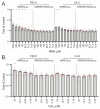
Poly(ADP-ribose) (PAR) is a molecular scaffold that aids in the formation of DNA repair protein complexes. Tools to sensitively quantify PAR in live cells have been lacking. We recently described the LivePAR probe (EGFP fused to the RNF146-encoded WWE PAR binding domain) to measure PAR formation at sites of laser micro-irradiation in live cells. Here, we present two methods that expand on the use of LivePAR and its WWE domain. First, LivePAR enriches in the nucleus of cells following genotoxic challenge. Image quantitation can identify single-cell PAR formation following genotoxic stress at concentrations lower than PAR ELISA or PAR immunoblot, with greater sensitivity to genotoxic stress than CometChip. In a second approach, we used the RNF146-encoded WWE domain to develop a split luciferase probe for analysis in a 96-well plate assay. We then applied these PAR analysis tools to demonstrate their broad applicability. First, we show that both approaches can identify genetic modifications that alter PARylation levels, such as hyper-PARylation in BRCA2-deficient cancer cells. Second, we demonstrate the utility of the WWE split luciferase assay to characterize the cellular response of genotoxins, PARP inhibitors, and PARG inhibitors, thereby providing a screening method to identify PAR modulating compounds.
Keywords: BER; BRCA2; DNA damage; LivePAR; PAR; Poly(ADP-ribose); WWE; split luciferase.
Conflict of interest statement
RWS is a scientific consultant for Canal House Biosciences, LLC but this company was not involved in nor was the consulting work related to this study. The authors declare no conflict of interest.
Figures




Similar articles
-
Overexpression of the WWE domain of RNF146 modulates poly-(ADP)-ribose dynamics at sites of DNA damage.bioRxiv [Preprint]. 2023 Dec 29:2023.12.29.573650. doi: 10.1101/2023.12.29.573650. bioRxiv. 2023. Update in: DNA Repair (Amst). 2025 Jun;150:103845. doi: 10.1016/j.dnarep.2025.103845. PMID: 38234836 Free PMC article. Updated. Preprint.
-
Overexpression of the WWE domain of RNF146 modulates poly-(ADP)-ribose dynamics at sites of DNA damage.DNA Repair (Amst). 2025 Jun;150:103845. doi: 10.1016/j.dnarep.2025.103845. Epub 2025 May 21. DNA Repair (Amst). 2025. PMID: 40403420 Free PMC article.
-
Quantitative Analysis of Nuclear Poly(ADP-Ribose) Dynamics in Response to Laser-Induced DNA Damage.Methods Mol Biol. 2023;2609:43-59. doi: 10.1007/978-1-0716-2891-1_3. Methods Mol Biol. 2023. PMID: 36515828 Free PMC article.
-
Poly-ADP-ribosylation dynamics, signaling, and analysis.Environ Mol Mutagen. 2024 Nov;65(9):315-337. doi: 10.1002/em.22623. Epub 2024 Sep 2. Environ Mol Mutagen. 2024. PMID: 39221603 Review.
-
The Molecular Mechanisms of Actions, Effects, and Clinical Implications of PARP Inhibitors in Epithelial Ovarian Cancers: A Systematic Review.Int J Mol Sci. 2022 Jul 23;23(15):8125. doi: 10.3390/ijms23158125. Int J Mol Sci. 2022. PMID: 35897700 Free PMC article.
Cited by
-
Overexpression of the WWE domain of RNF146 modulates poly-(ADP)-ribose dynamics at sites of DNA damage.bioRxiv [Preprint]. 2023 Dec 29:2023.12.29.573650. doi: 10.1101/2023.12.29.573650. bioRxiv. 2023. Update in: DNA Repair (Amst). 2025 Jun;150:103845. doi: 10.1016/j.dnarep.2025.103845. PMID: 38234836 Free PMC article. Updated. Preprint.
-
Novel In Vivo CometChip Reveals NDMA-Induced DNA Damage and Repair in Multiple Mouse Tissues.Int J Mol Sci. 2022 Oct 4;23(19):11776. doi: 10.3390/ijms231911776. Int J Mol Sci. 2022. PMID: 36233095 Free PMC article.
-
Overexpression of the WWE domain of RNF146 modulates poly-(ADP)-ribose dynamics at sites of DNA damage.DNA Repair (Amst). 2025 Jun;150:103845. doi: 10.1016/j.dnarep.2025.103845. Epub 2025 May 21. DNA Repair (Amst). 2025. PMID: 40403420 Free PMC article.
-
DNA polymerase beta expression in head & neck cancer modulates the poly(ADP-ribose)-mediated replication checkpoint.DNA Repair (Amst). 2025 Jun;150:103853. doi: 10.1016/j.dnarep.2025.103853. Epub 2025 Jun 2. DNA Repair (Amst). 2025. PMID: 40472742 Free PMC article.
-
Mechanisms of PARP1 inhibitor resistance and their implications for cancer treatment.NAR Cancer. 2022 Dec 22;4(4):zcac042. doi: 10.1093/narcan/zcac042. eCollection 2022 Dec. NAR Cancer. 2022. PMID: 36568963 Free PMC article.
References
-
- Koczor C.A., Saville K.M., Andrews J.F., Clark J., Fang Q., Li J., Al-Rahahleh R.Q., Ibrahim M., McClellan S., Makarov M.V., et al. Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD(+)/SIRT6 axis. Cell Rep. 2021;37:109917. doi: 10.1016/j.celrep.2021.109917. - DOI - PMC - PubMed
Grants and funding
- R44 ES032522/ES/NIEHS NIH HHS/United States
- R01 CA238061/CA/NCI NIH HHS/United States
- P01 ES028949/ES/NIEHS NIH HHS/United States
- R01 CA236911/CA/NCI NIH HHS/United States
- U01 ES029518/ES/NIEHS NIH HHS/United States
- GRANT11998991, DURIP-Navy/United States Department of Defense
- NSF-1841811/National Science Foundation
- R01 AG069740/AG/NIA NIH HHS/United States
- R01 CA148629/CA/NCI NIH HHS/United States
- ES029518, CA148629, ES014811, ES028949, CA238061, CA236911, AG069740 and ES032522/NH/NIH HHS/United States
- R01 ES014811/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

