Exosomes from EGFR-Mutated Adenocarcinoma Induce a Hybrid EMT and MMP9-Dependant Tumor Invasion
- PMID: 35954442
- PMCID: PMC9367273
- DOI: 10.3390/cancers14153776
Exosomes from EGFR-Mutated Adenocarcinoma Induce a Hybrid EMT and MMP9-Dependant Tumor Invasion
Abstract
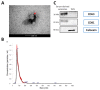
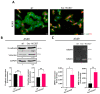
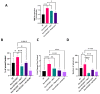
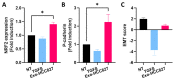
Exosomes, a class of extra cellular nano-sized vesicles (EVs), and their contents have gained attention as potential sources of information on tumor detection and regulatory drivers of tumor progression and metastasis. The effect of exosomes isolated from patients with an Epidermal Growth Factor Receptor (EGFR)-mutated adenocarcinoma on the promotion of epithelial-mesenchymal transition (EMT) and invasion were examined. Exosomes derived from serum of patients with EGFR-mutated non-small cell lung cancer (NSCLC) mediate the activation of the Phosphoinositide 3-kinase (PI3K)/AKT/ mammalian target of rapamycin (mTOR) pathway and induce an invasion through the up-regulation of matrix metalloproteinase-9 (MMP-9) in A549 cells. We observed a significant increase in the expression of vimentin, a mesenchymal marker, while retaining the epithelial characteristics, as evidenced by the unaltered levels of E-cadherin and Epithelial cell adhesion molecule (EPCAM). We also observed an increase of nuclear factor erythroid 2-related factor 2 (NFR2) and P-cadherin expression, markers of hybrid EMT. Exosomes derived from EGFR-mutated adenocarcinoma serum could be a potential mediator of hybrid EMT and tumor invasion. Understanding how cancerous cells communicate and interact with their environment via exosomes will improve our understanding of lung cancer progression and metastasis formation.
Keywords: EGFR; exosome; lung cancer; partial EMT.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

