BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition
- PMID: 35955412
- PMCID: PMC9368835
- DOI: 10.3390/ijms23158276
BPA Decreases PDCD4 in Bovine Granulosa Cells Independently of miR-21 Inhibition
Abstract
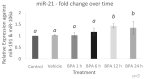
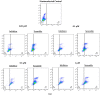
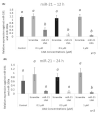
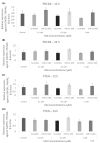
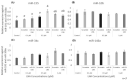
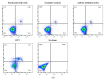
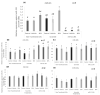
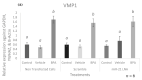
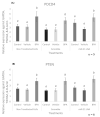
microRNAs (miRNAs) are susceptible to environmental factors that might affect cellular function and impose negative effects on female reproduction. miR-21 is the most abundant miRNA in bovine granulosa cells and is widely reported as affected by Bisphenol A (BPA) exposure, yet the cause and consequences are not entirely elucidated. BPA is a synthetic endocrine disruptor associated with poor fertility. miR-21 function in bovine granulosa cells is investigated utilizing locked nucleic acid (LNA) oligonucleotides to suppress miR-21. Before measuring apoptosis and quantifying miR-21 apoptotic targets PDCD4 and PTEN, transfection was optimized and validated. BPA was introduced to see how it affects miR-21 regulation and which BPA-mediated effects are influenced by miR-21. miR-21 knockdown and specificity against additional miRNAs were confirmed. miR-21 was found to have antiapoptotic effects, which could be explained by its effect on the proapoptotic target PDCD4, but not PTEN. Previous findings of miR-21 overexpression were validated using BPA treatments, and the temporal influence of BPA on miR-21 levels was addressed. Finally, BPA effects on upstream regulators, such as VMP1 and STAT3, explain the BPA-dependent upregulation of miR-21 expression. Overall, this research enhances our understanding of miR-21 function in granulosa cells and the mechanisms of BPA-induced reproductive impairment.
Keywords: BPA; PDCD4; PTEN; apoptosis; granulosa cells; miR-21.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

