Histone Marks-Dependent Effect on Alternative Splicing: New Perspectives for Targeted Splicing Modulation in Cancer?
- PMID: 35955433
- PMCID: PMC9368390
- DOI: 10.3390/ijms23158304
Histone Marks-Dependent Effect on Alternative Splicing: New Perspectives for Targeted Splicing Modulation in Cancer?
Abstract
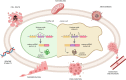
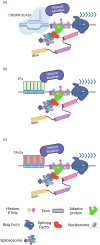
Alternative splicing (AS) is a tightly regulated mechanism that generates the complex human proteome from a small number of genes. Cis-regulatory RNA motifs in exons and introns control AS, recruiting positive and negative trans-acting splicing regulators. At a higher level, chromatin affects splicing events. Growing evidence indicates that the popular histone code hypothesis can be extended to RNA-level processes, such as AS. In addition to nucleosome positioning, which can generate transcriptional barriers to shape the final splicing outcome, histone post-translational modifications can contribute to the detailed regulation of single exon inclusion/exclusion. A histone-based system can identify alternatively spliced chromatin stretches, affecting RNAPII elongation locally or recruiting splicing components via adaptor complexes. In tumor cells, several mechanisms trigger misregulated AS events and produce cancer-associated transcripts. On a genome-wide level, aberrant AS can be the consequence of dysfunctional epigenetic splicing code, including altered enrichment in histone post-translational modifications. This review describes the main findings related to the effect of histone modifications and variants on splicing outcome and how a dysfunctional epigenetic splicing code triggers aberrant AS in cancer. In addition, it highlights recent advances in programmable DNA-targeting technologies and their possible application for AS targeted epigenetic modulation.
Keywords: alternative splicing; cancer transcript variants; epigenome editing; histone post-translational modifications; histone-code.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Splicing-associated chromatin signatures: a combinatorial and position-dependent role for histone marks in splicing definition.Nat Commun. 2021 Jan 29;12(1):682. doi: 10.1038/s41467-021-20979-x. Nat Commun. 2021. PMID: 33514745 Free PMC article.
-
Inferring causal relationships among histone modifications in exon skipping event.Methods. 2024 Dec;232:89-95. doi: 10.1016/j.ymeth.2024.11.008. Epub 2024 Nov 9. Methods. 2024. PMID: 39528091
-
Histone marks regulate the epithelial-to-mesenchymal transition via alternative splicing.Cell Rep. 2022 Feb 15;38(7):110357. doi: 10.1016/j.celrep.2022.110357. Cell Rep. 2022. PMID: 35172149
-
A saga of cancer epigenetics: linking epigenetics to alternative splicing.Biochem J. 2017 Mar 7;474(6):885-896. doi: 10.1042/BCJ20161047. Biochem J. 2017. PMID: 28270561 Review.
-
Integrative Chemical Biology Approaches to Deciphering the Histone Code: A Problem-Driven Journey.Acc Chem Res. 2021 Oct 5;54(19):3734-3747. doi: 10.1021/acs.accounts.1c00463. Epub 2021 Sep 23. Acc Chem Res. 2021. PMID: 34553920 Review.
Cited by
-
Regulation of Pre-mRNA Splicing: Indispensable Role of Post-Translational Modifications of Splicing Factors.Life (Basel). 2023 Feb 21;13(3):604. doi: 10.3390/life13030604. Life (Basel). 2023. PMID: 36983760 Free PMC article. Review.
-
Next-generation CRISPR technology for genome, epigenome and mitochondrial editing.Transgenic Res. 2024 Oct;33(5):323-357. doi: 10.1007/s11248-024-00404-x. Epub 2024 Aug 19. Transgenic Res. 2024. PMID: 39158822 Review.
-
Alternative splicing of KLF4 in myeloid cells: implications for cellular plasticity and trained immunity in cancer and inflammatory disease.Front Immunol. 2025 Jun 9;16:1585528. doi: 10.3389/fimmu.2025.1585528. eCollection 2025. Front Immunol. 2025. PMID: 40552304 Free PMC article. Review.
-
The arginine methyltransferase Prmt1 coordinates the germline arginine methylome essential for spermatogonial homeostasis and male fertility.Nucleic Acids Res. 2023 Oct 27;51(19):10428-10450. doi: 10.1093/nar/gkad769. Nucleic Acids Res. 2023. PMID: 37739418 Free PMC article.
-
LncRNA HOTAIRM1 promotes radioresistance in nasopharyngeal carcinoma by modulating FTO acetylation-dependent alternative splicing of CD44.Neoplasia. 2024 Oct;56:101034. doi: 10.1016/j.neo.2024.101034. Epub 2024 Aug 10. Neoplasia. 2024. PMID: 39128424 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

