Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis
- PMID: 35955502
- PMCID: PMC9369074
- DOI: 10.3390/ijms23158369
Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis
Erratum in
-
Correction: Mei et al. Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis. Int. J. Mol. Sci. 2022, 23, 8369.Int J Mol Sci. 2023 Jan 22;24(3):2224. doi: 10.3390/ijms24032224. Int J Mol Sci. 2023. PMID: 36769381 Free PMC article.
Abstract
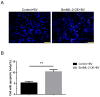
C-type lectins (CTLs) are widely distributed in mammals, insects, and plants, which act as pattern recognition receptors (PRRs) to recognize pathogens and initiate immune responses. In this study, we identified a C-type lectin gene called BmIML-2 from the silkworm Bombyx mori. Its open reading frame (ORF) encodes 314 amino acids, which contain dual tandem C-type lectin-like domain (CTLD). BmIML-2 is highly expressed in the fat body and is significantly induced at 24 h after BmNPV infection. Moreover, overexpression of BmIML-2 dramatically inhibited the proliferation of BmNPV, and knockdown assay via siRNA further validated the inhibition of BmIML-2 on viral proliferation. In addition, transcript level detection of apoptosis-related genes and observation of apoptosis bodies implied that overexpression of BmIML-2 promoted BmNPV-induced apoptosis. Immunofluorescence analysis indicated that BmIML-2 distributed throughout the cytoplasm and was slightly concentrated in the cell membrane. Taken together, our results suggest that BmIML-2 could inhibit in the proliferation of BmNPV by facilitating cell apoptosis.
Keywords: B. mori nucleopolyhedrovirus; Bombyx mori; C-type lectin; apoptosis.
Conflict of interest statement
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Study on the Role of Cytc in Response to BmNPV Infection in Silkworm, Bombyx mori (Lepidoptera).Int J Mol Sci. 2019 Sep 4;20(18):4325. doi: 10.3390/ijms20184325. Int J Mol Sci. 2019. PMID: 31487808 Free PMC article.
-
Bombyx mori PAT4 gene inhibits BmNPV infection and replication through autophagy.J Invertebr Pathol. 2025 Feb;208:108235. doi: 10.1016/j.jip.2024.108235. Epub 2024 Nov 21. J Invertebr Pathol. 2025. PMID: 39580048
-
The Bombyx mori G protein β subunit 1 (BmGNβ1) gene inhibits BmNPV infection.J Invertebr Pathol. 2024 Jun;204:108097. doi: 10.1016/j.jip.2024.108097. Epub 2024 Mar 25. J Invertebr Pathol. 2024. PMID: 38537687
-
The digestive proteinase trypsin, alkaline A contributes to anti-BmNPV activity in silkworm (Bombyx mori).Dev Comp Immunol. 2021 Jun;119:104035. doi: 10.1016/j.dci.2021.104035. Epub 2021 Jan 31. Dev Comp Immunol. 2021. PMID: 33535067
-
Discovery of anti-viral molecules and their vital functions in Bombyx mori.J Invertebr Pathol. 2018 May;154:12-18. doi: 10.1016/j.jip.2018.02.012. Epub 2018 Feb 14. J Invertebr Pathol. 2018. PMID: 29453967 Review.
Cited by
-
The Bmtret1 Gene Family and Its Potential Role in Response to BmNPV Stress in Bombyx mori.Int J Mol Sci. 2023 Dec 28;25(1):402. doi: 10.3390/ijms25010402. Int J Mol Sci. 2023. PMID: 38203572 Free PMC article.
-
Correction: Mei et al. Bombyx mori C-Type Lectin (BmIML-2) Inhibits the Proliferation of B. mori Nucleopolyhedrovirus (BmNPV) through Involvement in Apoptosis. Int. J. Mol. Sci. 2022, 23, 8369.Int J Mol Sci. 2023 Jan 22;24(3):2224. doi: 10.3390/ijms24032224. Int J Mol Sci. 2023. PMID: 36769381 Free PMC article.
-
Immune functions of pattern recognition receptors in Lepidoptera.Front Immunol. 2023 Jun 16;14:1203061. doi: 10.3389/fimmu.2023.1203061. eCollection 2023. Front Immunol. 2023. PMID: 37398667 Free PMC article. Review.
-
Regulatory networks of mRNAs and miRNAs involved in the immune response of diamondback moth, Plutella xylostella to fungal infection.BMC Genomics. 2025 Jan 7;26(1):15. doi: 10.1186/s12864-024-11192-3. BMC Genomics. 2025. PMID: 39762741 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

